| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
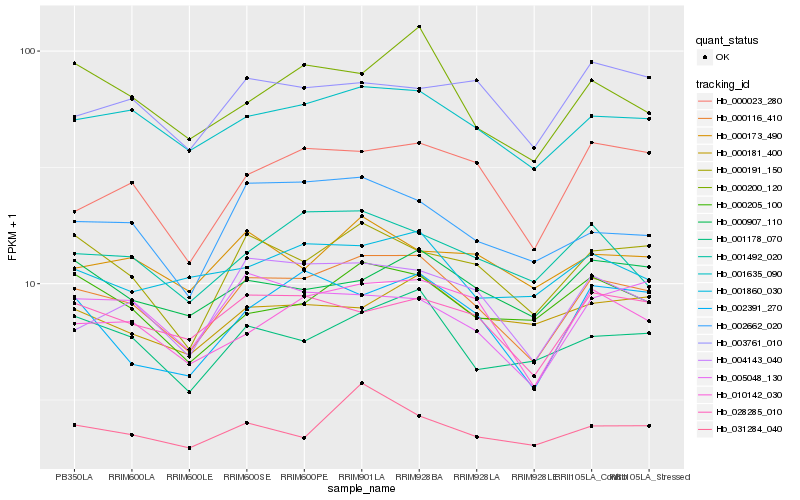
Hb_000116_410 |
0.0 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 2 |
Hb_001635_090 |
0.063895076 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 3 |
Hb_010142_030 |
0.0723774604 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000191_150 |
0.0732976656 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 5 |
Hb_004143_040 |
0.0771495113 |
- |
- |
argininosuccinate lyase, putative [Ricinus communis] |
| 6 |
Hb_001178_070 |
0.0785805555 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 7 |
Hb_028285_010 |
0.0797823327 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 8 |
Hb_002662_020 |
0.0813891951 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 9 |
Hb_000023_280 |
0.0834658762 |
- |
- |
hypothetical protein POPTR_0006s28210g [Populus trichocarpa] |
| 10 |
Hb_005048_130 |
0.0850716261 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 11 |
Hb_000200_120 |
0.0852408776 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000205_100 |
0.0859110295 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 13 |
Hb_002391_270 |
0.0885914186 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 14 |
Hb_001492_020 |
0.0893366978 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 15 |
Hb_000907_110 |
0.0902387763 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 16 |
Hb_031284_040 |
0.0908774933 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g30700 [Jatropha curcas] |
| 17 |
Hb_001860_030 |
0.0917568409 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 18 |
Hb_000173_490 |
0.0920502483 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000181_400 |
0.0926576426 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003761_010 |
0.0926636619 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |