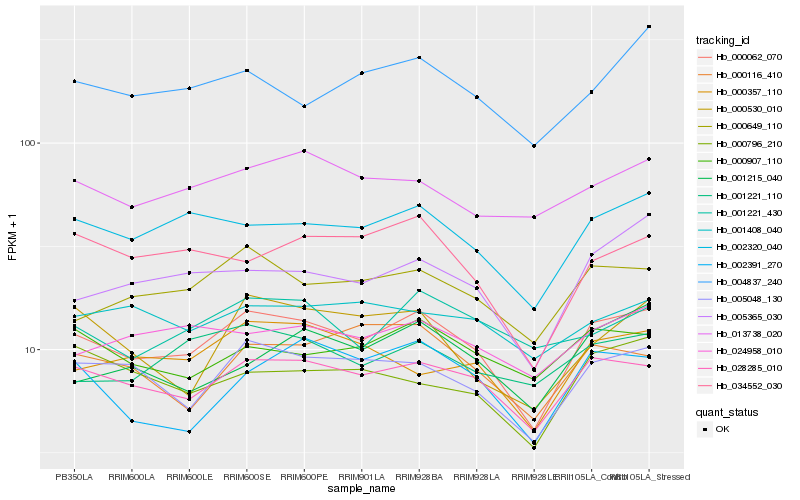
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_070 |
0.0 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 2 |
Hb_028285_010 |
0.0612152104 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 3 |
Hb_005048_130 |
0.0712114286 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 4 |
Hb_002320_040 |
0.0742357094 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 5 |
Hb_000649_110 |
0.0852584749 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 6 |
Hb_005365_030 |
0.0924508516 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 7 |
Hb_034552_030 |
0.0931386275 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_000357_110 |
0.0948274375 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 9 |
Hb_024958_010 |
0.0977432983 |
- |
- |
PREDICTED: LRR repeats and ubiquitin-like domain-containing protein At2g30105 [Jatropha curcas] |
| 10 |
Hb_013738_020 |
0.0984944903 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 11 |
Hb_000796_210 |
0.0994343256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000907_110 |
0.0997016541 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 13 |
Hb_000116_410 |
0.0997251306 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 14 |
Hb_004837_240 |
0.0998602847 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 15 |
Hb_002391_270 |
0.1006005411 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 16 |
Hb_001221_110 |
0.1019659107 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 17 |
Hb_001408_040 |
0.1020268623 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000530_010 |
0.1028779311 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |
| 19 |
Hb_001215_040 |
0.1036875619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001221_430 |
0.1043884285 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At5g63520 isoform X2 [Jatropha curcas] |