| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
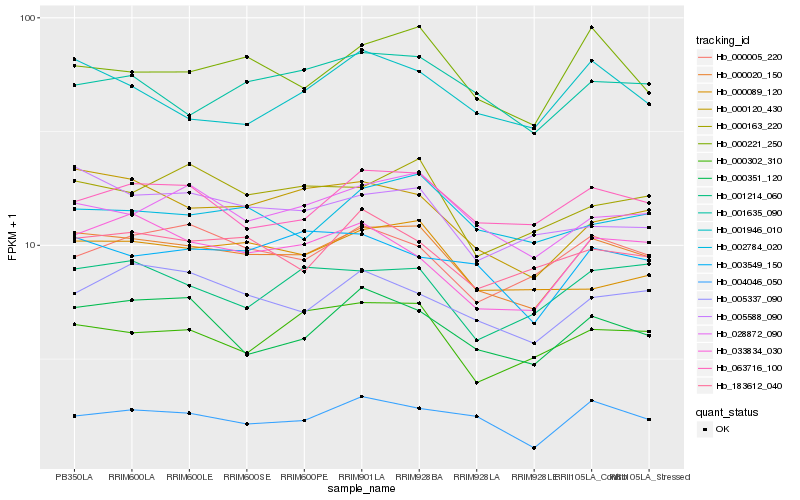
Hb_000020_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 2 |
Hb_028872_090 |
0.0659294645 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 3 |
Hb_000120_430 |
0.0670075031 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 4 |
Hb_063716_100 |
0.0694399668 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000221_250 |
0.0706618344 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |
| 6 |
Hb_033834_030 |
0.0706730226 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_183612_040 |
0.0733292002 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 8 |
Hb_000163_220 |
0.0733336893 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 9 |
Hb_005337_090 |
0.0734932038 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 10 |
Hb_000005_220 |
0.0736965876 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 11 |
Hb_000351_120 |
0.0742405703 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 12 |
Hb_002784_020 |
0.0751567991 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 13 |
Hb_005588_090 |
0.0753715445 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 14 |
Hb_003549_150 |
0.0753930066 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 15 |
Hb_004046_050 |
0.0759590711 |
- |
- |
PREDICTED: origin of replication complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_000302_310 |
0.0760955125 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 17 |
Hb_001214_060 |
0.0763543388 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC8 [Jatropha curcas] |
| 18 |
Hb_001635_090 |
0.0764124527 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 19 |
Hb_000089_120 |
0.0785147636 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_001946_010 |
0.0788141048 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |