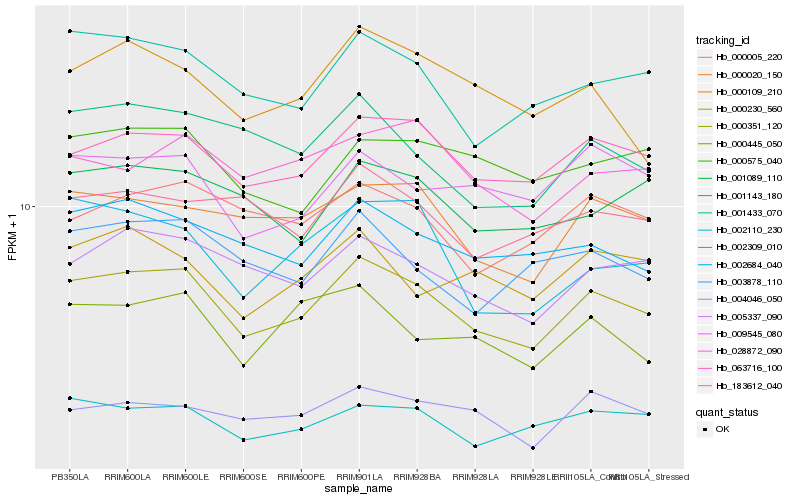
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000351_120 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 2 |
Hb_063716_100 |
0.0624404653 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001433_070 |
0.068748661 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 4 |
Hb_005337_090 |
0.0711427068 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 5 |
Hb_000020_150 |
0.0742405703 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 6 |
Hb_002684_040 |
0.0758736806 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002110_230 |
0.078865142 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 8 |
Hb_001143_180 |
0.0808485715 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 9 |
Hb_001089_110 |
0.0831720946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009545_080 |
0.0836682641 |
- |
- |
hypothetical protein POPTR_0008s15400g [Populus trichocarpa] |
| 11 |
Hb_000005_220 |
0.0836965957 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 12 |
Hb_003878_110 |
0.0837183987 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 13 |
Hb_000445_050 |
0.0849455219 |
- |
- |
PREDICTED: DNA topoisomerase 2-binding protein 1 [Jatropha curcas] |
| 14 |
Hb_000109_210 |
0.0854003046 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_002309_010 |
0.0855109805 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 16 |
Hb_004046_050 |
0.0856201161 |
- |
- |
PREDICTED: origin of replication complex subunit 3 [Jatropha curcas] |
| 17 |
Hb_000575_040 |
0.0864698933 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 18 |
Hb_183612_040 |
0.0872106355 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 19 |
Hb_028872_090 |
0.0873009651 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 20 |
Hb_000230_560 |
0.0881072769 |
- |
- |
PREDICTED: UPF0613 protein PB24D3.06c [Jatropha curcas] |