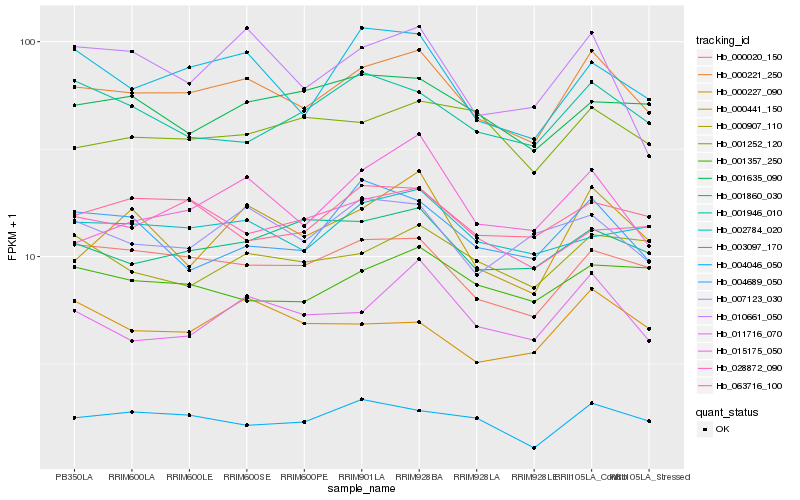
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000221_250 |
0.0 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |
| 2 |
Hb_000020_150 |
0.0706618344 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 3 |
Hb_003097_170 |
0.0801143239 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 4 |
Hb_011716_070 |
0.0817438011 |
transcription factor |
TF Family: HRT |
PREDICTED: uncharacterized protein LOC105637858 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001860_030 |
0.0843830249 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 6 |
Hb_004046_050 |
0.0865667817 |
- |
- |
PREDICTED: origin of replication complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_007123_030 |
0.0865922245 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 8 |
Hb_002784_020 |
0.0869647239 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 9 |
Hb_000441_150 |
0.0906579794 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_063716_100 |
0.0911059117 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001357_250 |
0.0915933976 |
- |
- |
PREDICTED: UPF0505 protein C16orf62 homolog [Jatropha curcas] |
| 12 |
Hb_000907_110 |
0.0929632593 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 13 |
Hb_010661_050 |
0.0938296548 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP]-like [Fragaria vesca subsp. vesca] |
| 14 |
Hb_004689_050 |
0.0940826696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001635_090 |
0.0940965954 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 16 |
Hb_001252_120 |
0.0946018382 |
- |
- |
PREDICTED: delta-1-pyrroline-5-carboxylate synthase-like [Gossypium raimondii] |
| 17 |
Hb_000227_090 |
0.0948092727 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 catalytic subunit [Jatropha curcas] |
| 18 |
Hb_015175_050 |
0.0952807636 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 19 |
Hb_001946_010 |
0.0960877323 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 20 |
Hb_028872_090 |
0.0967952546 |
- |
- |
PREDICTED: urease [Jatropha curcas] |