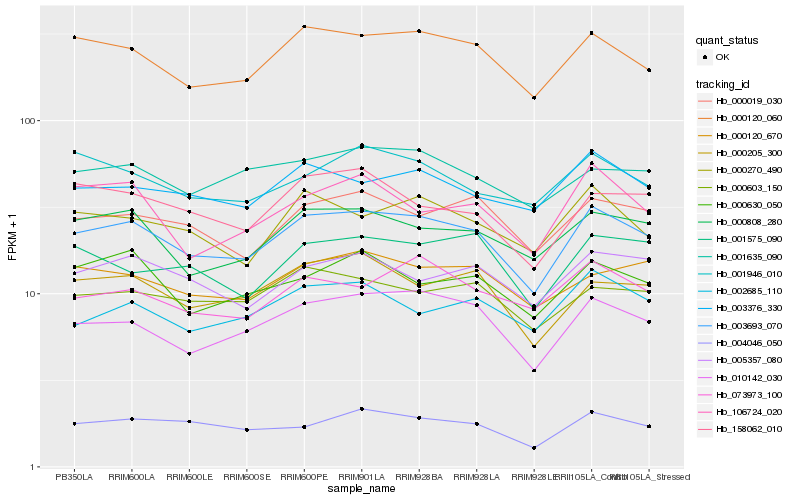
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003693_070 |
0.0 |
- |
- |
PREDICTED: S-(hydroxymethyl)glutathione dehydrogenase [Jatropha curcas] |
| 2 |
Hb_000120_060 |
0.054736454 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 3 |
Hb_010142_030 |
0.0576541002 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000808_280 |
0.0693630616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004046_050 |
0.0709736383 |
- |
- |
PREDICTED: origin of replication complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_000205_300 |
0.0711877334 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 7 |
Hb_000019_030 |
0.0720810301 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 8 |
Hb_003376_330 |
0.0751531475 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 9 |
Hb_000603_150 |
0.0755655052 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 10 |
Hb_000270_490 |
0.0769005731 |
- |
- |
betaine aldehyde dehydrogenase 1 [Jatropha curcas] |
| 11 |
Hb_001635_090 |
0.0771315456 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 12 |
Hb_005357_080 |
0.0774277956 |
- |
- |
PREDICTED: tyrosyl-DNA phosphodiesterase 1 [Jatropha curcas] |
| 13 |
Hb_158062_010 |
0.0785018165 |
- |
- |
PREDICTED: uncharacterized protein LOC105634413 [Jatropha curcas] |
| 14 |
Hb_000120_670 |
0.0787471666 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |
| 15 |
Hb_000630_050 |
0.0791545322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_073973_100 |
0.0817794604 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 17 |
Hb_001575_090 |
0.0823515896 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 18 |
Hb_001946_010 |
0.0827304343 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 19 |
Hb_002685_110 |
0.0834017003 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 20 |
Hb_106724_020 |
0.0837588736 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 1A-like isoform X1 [Populus euphratica] |