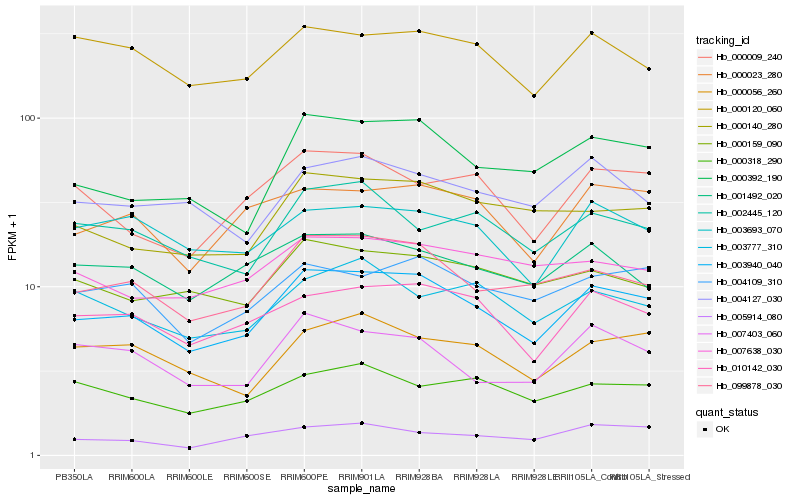
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003940_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_099878_030 |
0.0729802241 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 3 |
Hb_010142_030 |
0.0782599211 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000392_190 |
0.0849240739 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 5 |
Hb_004109_310 |
0.0870758841 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 6 |
Hb_000023_280 |
0.0887073771 |
- |
- |
hypothetical protein POPTR_0006s28210g [Populus trichocarpa] |
| 7 |
Hb_000140_280 |
0.0928719123 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 8 |
Hb_000056_260 |
0.0939912328 |
- |
- |
hypothetical protein CICLE_v10033822mg [Citrus clementina] |
| 9 |
Hb_007403_060 |
0.0972964083 |
- |
- |
PREDICTED: CAAX prenyl protease 2 [Jatropha curcas] |
| 10 |
Hb_003693_070 |
0.0977285223 |
- |
- |
PREDICTED: S-(hydroxymethyl)glutathione dehydrogenase [Jatropha curcas] |
| 11 |
Hb_005914_080 |
0.0985941679 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000009_240 |
0.0991610883 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000159_090 |
0.0995510933 |
- |
- |
hypothetical protein POPTR_0015s153201g, partial [Populus trichocarpa] |
| 14 |
Hb_000120_060 |
0.099936008 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 15 |
Hb_001492_020 |
0.1006477047 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 16 |
Hb_004127_030 |
0.1009911254 |
- |
- |
putative Rab geranylgeranyl transferase type II beta subunit family protein [Populus trichocarpa] |
| 17 |
Hb_002445_120 |
0.1012929776 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 18 |
Hb_000318_290 |
0.1028206067 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007638_030 |
0.1033115153 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 20 |
Hb_003777_310 |
0.1040082373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |