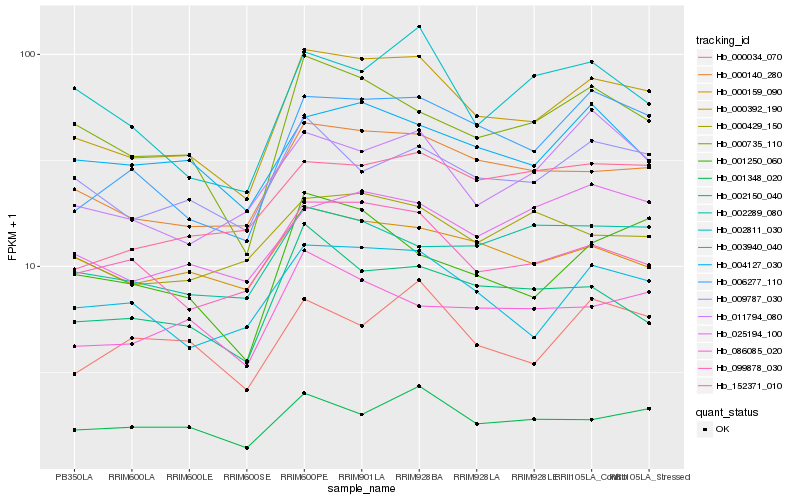
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_190 |
0.0 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 2 |
Hb_000140_280 |
0.0743412345 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 3 |
Hb_006277_110 |
0.0832685661 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_003940_040 |
0.0849240739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_099878_030 |
0.0912705091 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 6 |
Hb_000735_110 |
0.091808231 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |
| 7 |
Hb_009787_030 |
0.097345459 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 8 |
Hb_001348_020 |
0.0986045649 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001250_060 |
0.1038137976 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 10 |
Hb_025194_100 |
0.1041361846 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 11 |
Hb_086085_020 |
0.1045051265 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 12 |
Hb_000159_090 |
0.104828378 |
- |
- |
hypothetical protein POPTR_0015s153201g, partial [Populus trichocarpa] |
| 13 |
Hb_004127_030 |
0.1053859585 |
- |
- |
putative Rab geranylgeranyl transferase type II beta subunit family protein [Populus trichocarpa] |
| 14 |
Hb_011794_080 |
0.1057321474 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 15 |
Hb_002150_040 |
0.1060186218 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 16 |
Hb_002811_030 |
0.1067253146 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 17 |
Hb_000429_150 |
0.1101630773 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 18 |
Hb_000034_070 |
0.1105055696 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 3B-like [Jatropha curcas] |
| 19 |
Hb_002289_080 |
0.1115373933 |
- |
- |
hypothetical protein POPTR_0016s05590g [Populus trichocarpa] |
| 20 |
Hb_152371_010 |
0.1122235042 |
- |
- |
RNA-binding KH domain-containing protein [Theobroma cacao] |