| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
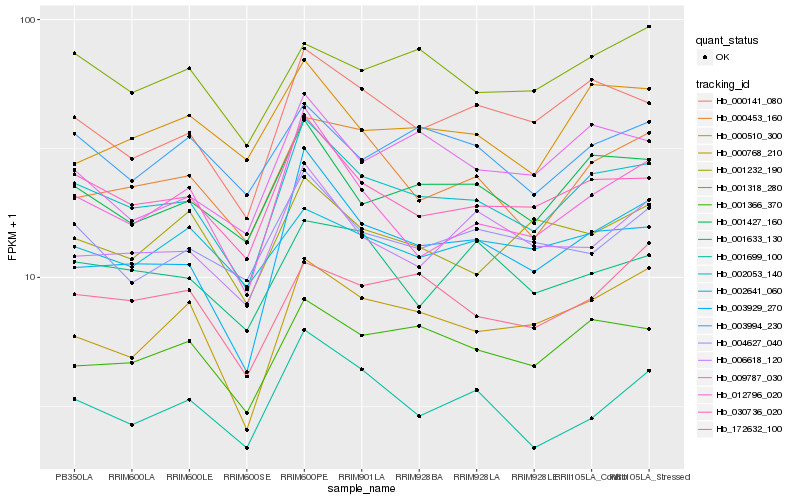
Hb_002053_140 |
0.0 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 2 |
Hb_030736_020 |
0.0497028976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001427_160 |
0.063800746 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000141_080 |
0.0639843031 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Brassica rapa] |
| 5 |
Hb_003929_270 |
0.0681475936 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001366_370 |
0.0683225454 |
- |
- |
PREDICTED: aarF domain-containing protein kinase 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_012796_020 |
0.0751982643 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |
| 8 |
Hb_000453_160 |
0.0758370723 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 9 |
Hb_000768_210 |
0.0820475194 |
- |
- |
PREDICTED: uncharacterized protein LOC105632522 [Jatropha curcas] |
| 10 |
Hb_004627_040 |
0.083579288 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 11 |
Hb_006618_120 |
0.0837362394 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 12 |
Hb_009787_030 |
0.0846874739 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 13 |
Hb_001699_100 |
0.0850859623 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 14 |
Hb_172632_100 |
0.0862457809 |
- |
- |
PREDICTED: reticulon-like protein B22 [Jatropha curcas] |
| 15 |
Hb_003994_230 |
0.086990678 |
- |
- |
7-dehydrocholesterol reductase, putative [Ricinus communis] |
| 16 |
Hb_001318_280 |
0.0894679135 |
- |
- |
Protein FAM18B, putative [Ricinus communis] |
| 17 |
Hb_001633_130 |
0.0902659517 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 18 |
Hb_000510_300 |
0.0906052489 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001232_190 |
0.0937282555 |
- |
- |
PREDICTED: uncharacterized protein LOC105639761 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002641_060 |
0.0937366118 |
- |
- |
prefoldin subunit, putative [Ricinus communis] |