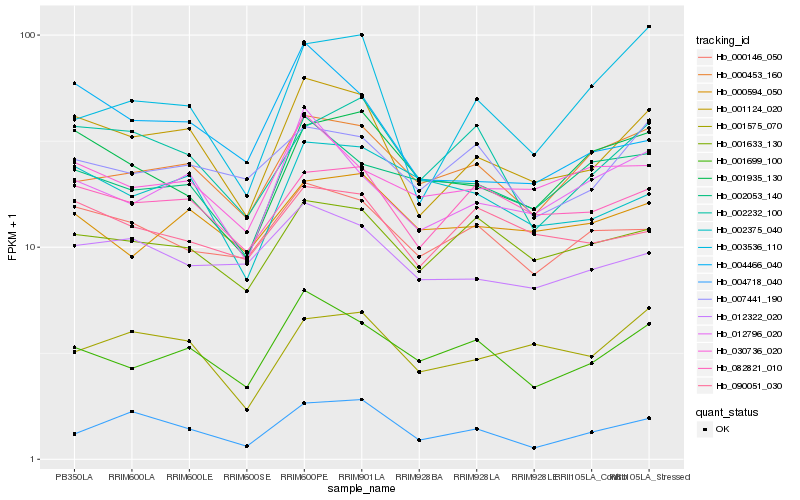
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001124_020 |
0.0 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 2 |
Hb_012796_020 |
0.0971026147 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |
| 3 |
Hb_082821_010 |
0.1012187302 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 4 |
Hb_001699_100 |
0.1013288643 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 5 |
Hb_001633_130 |
0.1087399768 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 6 |
Hb_001575_070 |
0.1088491038 |
- |
- |
PREDICTED: uncharacterized protein LOC105639885 [Jatropha curcas] |
| 7 |
Hb_004466_040 |
0.1118396342 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 8 |
Hb_000453_160 |
0.1128574935 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 9 |
Hb_000146_050 |
0.1131467271 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |
| 10 |
Hb_012322_020 |
0.1156232126 |
- |
- |
- |
| 11 |
Hb_002232_100 |
0.1161516556 |
- |
- |
vacuolar ATPase F subunit [Hevea brasiliensis] |
| 12 |
Hb_090051_030 |
0.1173514879 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 13 |
Hb_001935_130 |
0.1176311608 |
- |
- |
PREDICTED: uncharacterized protein LOC105643176 [Jatropha curcas] |
| 14 |
Hb_030736_020 |
0.1176738934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002375_040 |
0.1218254721 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 16 |
Hb_007441_190 |
0.1232587516 |
- |
- |
hypothetical protein CISIN_1g023046mg [Citrus sinensis] |
| 17 |
Hb_000594_050 |
0.1243865159 |
- |
- |
PREDICTED: pH-response regulator protein palA/RIM20 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002053_140 |
0.1245148706 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 19 |
Hb_004718_040 |
0.1253748373 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 20 |
Hb_003536_110 |
0.1255006983 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |