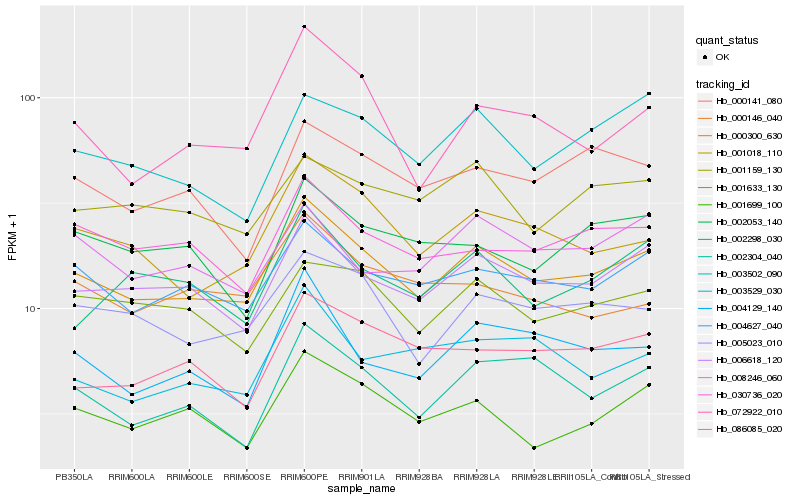
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_630 |
0.0 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 2 |
Hb_006618_120 |
0.0632317623 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 3 |
Hb_004627_040 |
0.0685686876 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 4 |
Hb_005023_010 |
0.0761183043 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 5 |
Hb_001018_110 |
0.0763491738 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_001699_100 |
0.0853291487 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 7 |
Hb_001633_130 |
0.0866085829 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 8 |
Hb_072922_010 |
0.0871044317 |
- |
- |
apolipoprotein d, putative [Ricinus communis] |
| 9 |
Hb_030736_020 |
0.087226515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004129_140 |
0.0925016956 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 11 |
Hb_000141_080 |
0.0927017932 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Brassica rapa] |
| 12 |
Hb_001159_130 |
0.0929539859 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 13 |
Hb_003502_090 |
0.0933142137 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 14 |
Hb_086085_020 |
0.0939648786 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 15 |
Hb_002304_040 |
0.0940172912 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002298_030 |
0.0953420137 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 17 |
Hb_008246_060 |
0.0968161351 |
- |
- |
hypothetical protein F383_01577 [Gossypium arboreum] |
| 18 |
Hb_000146_040 |
0.0970985857 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 19 |
Hb_003529_030 |
0.0983092059 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 20 |
Hb_002053_140 |
0.0985720736 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |