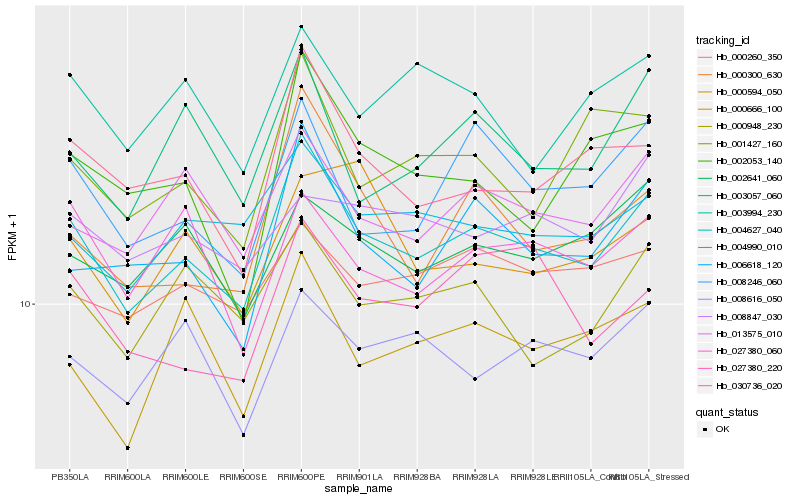
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004627_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 2 |
Hb_006618_120 |
0.0665136175 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 3 |
Hb_013575_010 |
0.0665363418 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 4 |
Hb_030736_020 |
0.0677441973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000300_630 |
0.0685686876 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 6 |
Hb_004990_010 |
0.069095873 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_008246_060 |
0.0706506915 |
- |
- |
hypothetical protein F383_01577 [Gossypium arboreum] |
| 8 |
Hb_000948_230 |
0.0743738588 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_003057_060 |
0.0747926169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002641_060 |
0.0759109817 |
- |
- |
prefoldin subunit, putative [Ricinus communis] |
| 11 |
Hb_000260_350 |
0.0764733441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001427_160 |
0.0781955068 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_008616_050 |
0.078477754 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 14 |
Hb_000666_100 |
0.0787223526 |
- |
- |
Actin-related protein 2/3 complex subunit 2 isoform 2 [Theobroma cacao] |
| 15 |
Hb_027380_220 |
0.0814704806 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 16 |
Hb_003994_230 |
0.0816802861 |
- |
- |
7-dehydrocholesterol reductase, putative [Ricinus communis] |
| 17 |
Hb_027380_060 |
0.0820884657 |
- |
- |
PREDICTED: putative ALA-interacting subunit 2 [Jatropha curcas] |
| 18 |
Hb_002053_140 |
0.083579288 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 19 |
Hb_008847_030 |
0.0864129587 |
- |
- |
PREDICTED: TLD domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_000594_050 |
0.0867594566 |
- |
- |
PREDICTED: pH-response regulator protein palA/RIM20 isoform X1 [Jatropha curcas] |