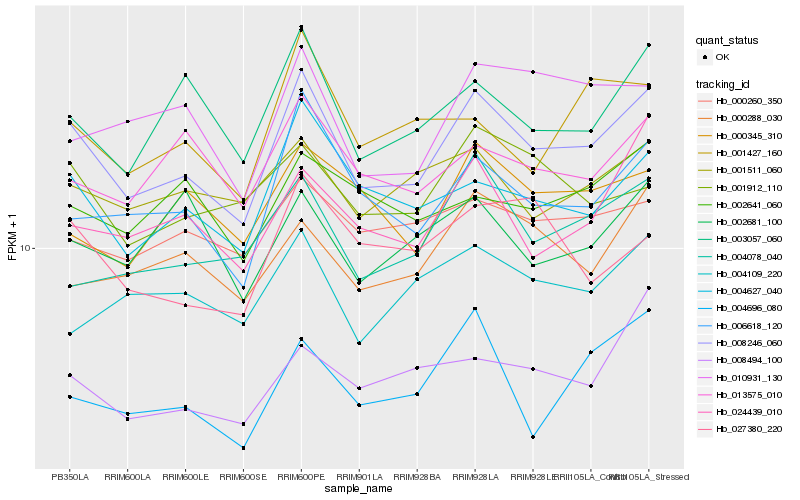
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008246_060 |
0.0 |
- |
- |
hypothetical protein F383_01577 [Gossypium arboreum] |
| 2 |
Hb_004627_040 |
0.0706506915 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 3 |
Hb_006618_120 |
0.0724074918 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 4 |
Hb_003057_060 |
0.0754076082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_024439_010 |
0.0824227305 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 6 |
Hb_013575_010 |
0.0836863345 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 7 |
Hb_001511_060 |
0.0837751684 |
- |
- |
PREDICTED: sorting and assembly machinery component 50 homolog [Jatropha curcas] |
| 8 |
Hb_000288_030 |
0.0848124088 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 9 |
Hb_000260_350 |
0.0851158603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004078_040 |
0.0857096637 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 11 |
Hb_000345_310 |
0.0871901188 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein isoform X3 [Jatropha curcas] |
| 12 |
Hb_001912_110 |
0.0880454556 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_010931_130 |
0.0880751126 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 14 |
Hb_001427_160 |
0.0885605829 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_008494_100 |
0.0889123344 |
- |
- |
PREDICTED: protein RMD5 homolog A-like [Jatropha curcas] |
| 16 |
Hb_004109_220 |
0.0901303403 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 9-like [Populus euphratica] |
| 17 |
Hb_004696_080 |
0.0901817758 |
- |
- |
PREDICTED: uncharacterized protein At1g01500-like [Populus euphratica] |
| 18 |
Hb_002641_060 |
0.0903732298 |
- |
- |
prefoldin subunit, putative [Ricinus communis] |
| 19 |
Hb_027380_220 |
0.0904559945 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 20 |
Hb_002681_100 |
0.0906482492 |
- |
- |
PREDICTED: glucuronokinase 1 [Jatropha curcas] |