| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
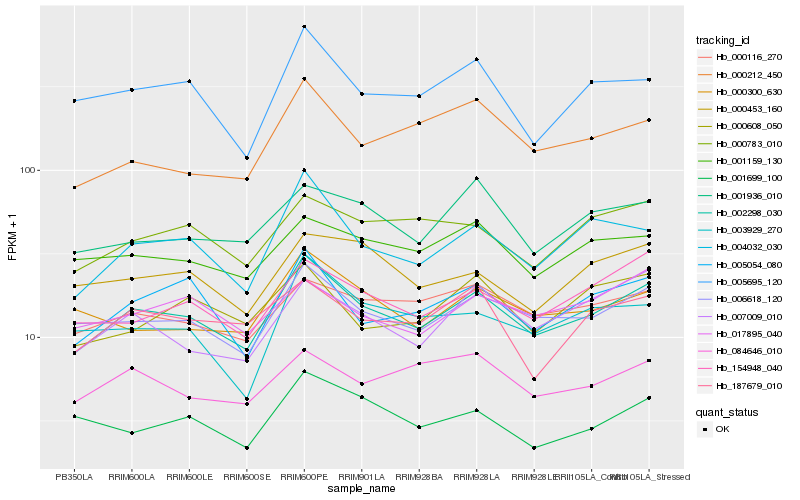
Hb_002298_030 |
0.0 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 2 |
Hb_006618_120 |
0.0663158897 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 3 |
Hb_004032_030 |
0.0782530605 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 4 |
Hb_005054_080 |
0.0835768971 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 5 |
Hb_005695_120 |
0.0907702201 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 6 |
Hb_154948_040 |
0.0911332938 |
- |
- |
hypothetical protein B456_013G043800 [Gossypium raimondii] |
| 7 |
Hb_000300_630 |
0.0953420137 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 8 |
Hb_000116_270 |
0.095367124 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |
| 9 |
Hb_000783_010 |
0.0954394219 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 10 |
Hb_001159_130 |
0.0980300817 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 11 |
Hb_000453_160 |
0.0986140347 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 12 |
Hb_187679_010 |
0.0989006125 |
- |
- |
- |
| 13 |
Hb_000212_450 |
0.0993372193 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 14 |
Hb_007009_010 |
0.0996336997 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675 [Populus euphratica] |
| 15 |
Hb_001936_010 |
0.0996873272 |
- |
- |
hypothetical protein Csa_2G348280 [Cucumis sativus] |
| 16 |
Hb_003929_270 |
0.1007011116 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_017895_040 |
0.1012558968 |
- |
- |
hypothetical protein POPTR_0006s08120g [Populus trichocarpa] |
| 18 |
Hb_000608_050 |
0.1017844809 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |
| 19 |
Hb_001699_100 |
0.1024363318 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 20 |
Hb_084646_010 |
0.1032854022 |
- |
- |
hypothetical protein CICLE_v10031746mg [Citrus clementina] |