| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
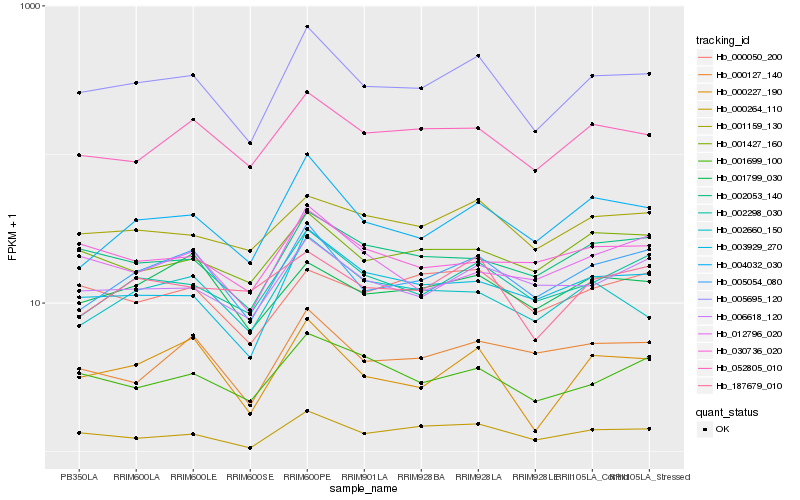
Hb_005695_120 |
0.0 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 2 |
Hb_005054_080 |
0.0852068485 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 3 |
Hb_002298_030 |
0.0907702201 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 4 |
Hb_052805_010 |
0.0921194762 |
- |
- |
hypothetical protein POPTR_0009s03650g [Populus trichocarpa] |
| 5 |
Hb_004032_030 |
0.0922592452 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 6 |
Hb_003929_270 |
0.0932272809 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000264_110 |
0.0954833882 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 13 [Jatropha curcas] |
| 8 |
Hb_002053_140 |
0.0983272236 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 9 |
Hb_006618_120 |
0.0993239862 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 10 |
Hb_001159_130 |
0.1017846648 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 11 |
Hb_001699_100 |
0.1022770931 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 12 |
Hb_001427_160 |
0.1079218871 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_002660_150 |
0.1119188525 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 14 |
Hb_187679_010 |
0.1123565107 |
- |
- |
- |
| 15 |
Hb_001799_030 |
0.1143712459 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 16 |
Hb_000050_200 |
0.1152527774 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 17 |
Hb_030736_020 |
0.1152692999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000227_190 |
0.1154471066 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000127_140 |
0.1158594984 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 20 |
Hb_012796_020 |
0.1158909727 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |