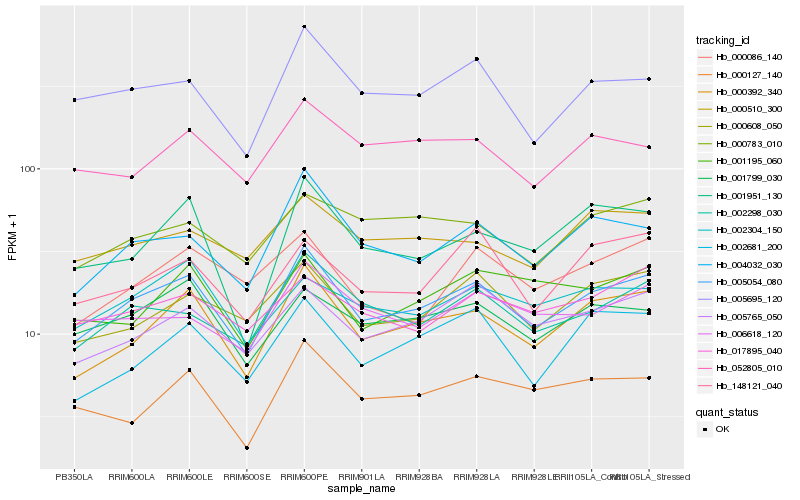
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_080 |
0.0 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 2 |
Hb_000392_340 |
0.0619272204 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 3 |
Hb_004032_030 |
0.0826492999 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 4 |
Hb_002298_030 |
0.0835768971 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 5 |
Hb_005695_120 |
0.0852068485 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 6 |
Hb_000608_050 |
0.0923045679 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |
| 7 |
Hb_005765_050 |
0.0931154582 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4-like [Populus euphratica] |
| 8 |
Hb_001799_030 |
0.0931403213 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 9 |
Hb_001951_130 |
0.0985717681 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 10 |
Hb_002681_200 |
0.0986030444 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 11 |
Hb_000127_140 |
0.0988876636 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 12 |
Hb_000783_010 |
0.1023168323 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 13 |
Hb_052805_010 |
0.1024100059 |
- |
- |
hypothetical protein POPTR_0009s03650g [Populus trichocarpa] |
| 14 |
Hb_000510_300 |
0.1026420837 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_002304_150 |
0.1033254913 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001195_060 |
0.1036056955 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 17 |
Hb_006618_120 |
0.1049815758 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 18 |
Hb_017895_040 |
0.1056309505 |
- |
- |
hypothetical protein POPTR_0006s08120g [Populus trichocarpa] |
| 19 |
Hb_148121_040 |
0.1066693595 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000086_140 |
0.1071336688 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |