| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
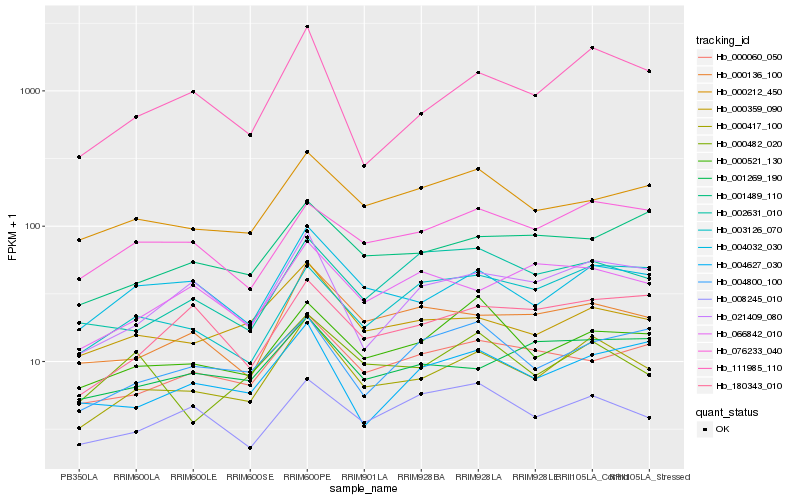
Hb_000417_100 |
0.0 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 2 |
Hb_004032_030 |
0.1001563842 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 3 |
Hb_111985_110 |
0.1056790251 |
- |
- |
thioredoxin H-type 3 [Hevea brasiliensis] |
| 4 |
Hb_000359_090 |
0.112675183 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 5 |
Hb_000136_100 |
0.1139129386 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 6 |
Hb_021409_080 |
0.1145377593 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 7 |
Hb_000521_130 |
0.1223922555 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_076233_040 |
0.123637168 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000212_450 |
0.1278464635 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 10 |
Hb_000060_050 |
0.1297608479 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 11 |
Hb_000482_020 |
0.1313892538 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 12 |
Hb_001269_190 |
0.1328118539 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 13 |
Hb_008245_010 |
0.1350588845 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 14 |
Hb_002631_010 |
0.136775188 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 15 |
Hb_180343_010 |
0.1378472942 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 16 |
Hb_004627_030 |
0.137957351 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 17 |
Hb_001489_110 |
0.1389627823 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |
| 18 |
Hb_066842_010 |
0.1393383795 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003126_070 |
0.1393518786 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004800_100 |
0.1399189916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |