Hb_111985_110
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig111985: 70985-72612 |
| Sequence |   |
Annotation
kegg
| ID | rcu:RCOM_1595970 |
|---|---|
| description | Thioredoxin H-type, putative |
nr
| ID | AEC03317.1 |
|---|---|
| description | thioredoxin H-type 3 [Hevea brasiliensis] |
swissprot
| ID | Q43636 |
|---|---|
| description | Thioredoxin H-type OS=Ricinus communis PE=3 SV=1 |
trembl
| ID | I1SSI8 |
|---|---|
| description | Thioredoxin H-type 3 OS=Hevea brasiliensis GN=TrxH3 PE=2 SV=1 |
Gene Ontology
| ID | GO:0005829 |
|---|---|
| description | thioredoxin h-type |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_03618: 70673-72681 |
|---|---|
| cDNA (Sanger) (ID:Location) |
001_L01.ab1: 70814-72681 , 004_B05.ab1: 70775-72678 , 004_F23.ab1: 70795-72684 , 006_E11.ab1: 70876-72678 , 006_I24.ab1: 70780-72681 , 007_L08.ab1: 70876-72678 , 008_I19.ab1: 70875-72678 , 008_J04.ab1: 70877-72678 , 009_J14.ab1: 70770-72681 , 009_L14.ab1: 70876-72681 , 018_E23.ab1: 70798-72618 , 018_H02.ab1: 70897-72681 , 021_A12.ab1: 70872-72681 , 023_I19.ab1: 70937-72681 , 023_I21.ab1: 70878-72681 , 024_D17.ab1: 70880-72678 , 024_J16.ab1: 70876-72682 , 026_M11.ab1: 70885-72676 , 031_N18.ab1: 70775-72678 , 033_B20.ab1: 70876-72678 , 035_B17.ab1: 70781-72681 , 035_D19.ab1: 70812-72681 , 036_J04.ab1: 70798-72684 , 038_O01.ab1: 70800-72684 , 039_H19.ab1: 70876-72681 , 041_E17.ab1: 70800-72684 , 041_O15.ab1: 70814-72681 , 043_D01.ab1: 70798-72681 , 048_G09.ab1: 70781-72681 , 049_F06.ab1: 70798-72678 , 050_E23.ab1: 70876-72678 , 052_O11.ab1: 70900-72681 |
Similar expressed genes (Top20)
Gene co-expression network
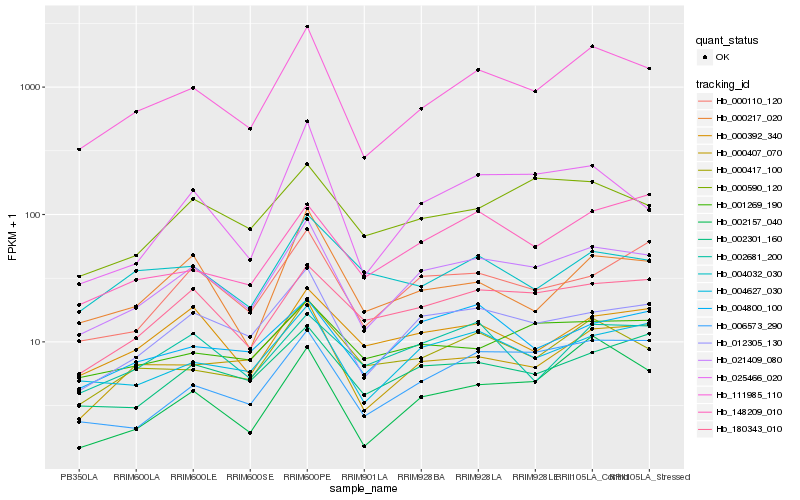
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 640.556 | 470.895 | 986.68 | 2986.58 | 323.275 | 278.615 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 2085.1 | 1397.55 | 1367.25 | 677.12 | 924.308 |

