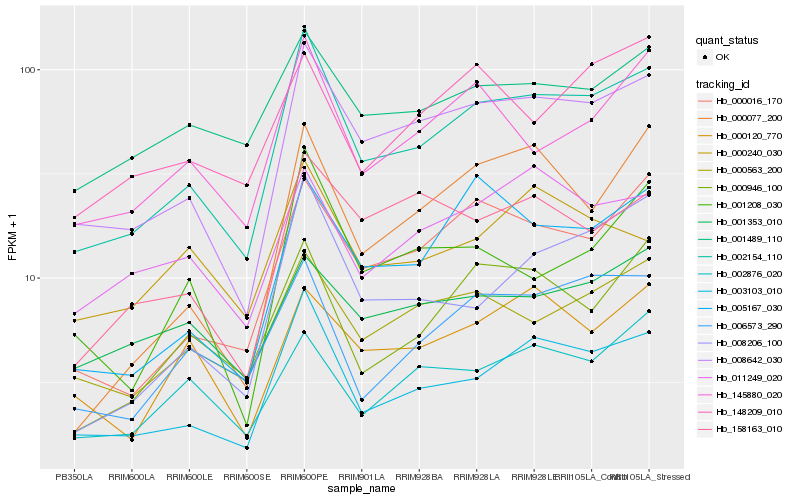
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002154_110 |
0.0 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 2 |
Hb_003103_010 |
0.065594896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008642_030 |
0.0715805713 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000016_170 |
0.1121931503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000563_200 |
0.1152637641 |
- |
- |
hypothetical protein JCGZ_18930 [Jatropha curcas] |
| 6 |
Hb_006573_290 |
0.1160090926 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 7 |
Hb_145880_020 |
0.1171245362 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 8 |
Hb_005167_030 |
0.1208226998 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 9 |
Hb_008206_100 |
0.1211269944 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 10 |
Hb_000946_100 |
0.1214863804 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 11 |
Hb_001489_110 |
0.1311104823 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |
| 12 |
Hb_001208_030 |
0.1333868708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011249_020 |
0.1337780736 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 14 |
Hb_158163_010 |
0.1368735474 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_002876_020 |
0.1402357077 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000120_770 |
0.1408712947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001353_010 |
0.1416448517 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX11, mitochondrial [Jatropha curcas] |
| 18 |
Hb_148209_010 |
0.1433748266 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 19 |
Hb_000077_200 |
0.1450453848 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 20 |
Hb_000240_030 |
0.1450577513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |