| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
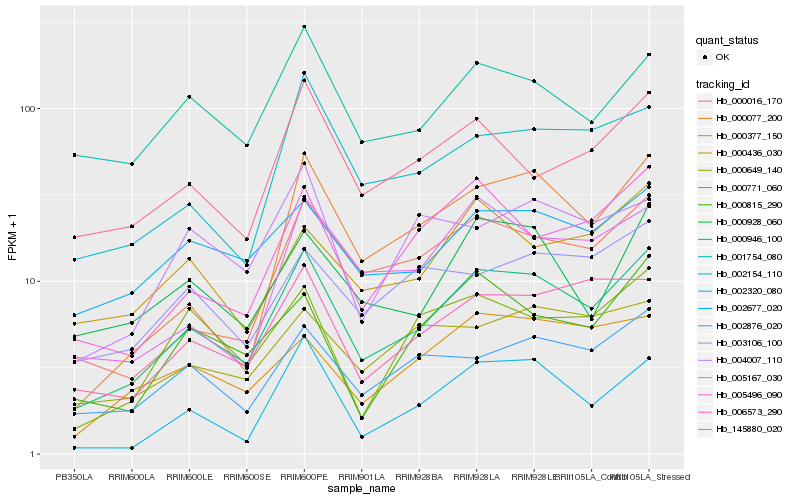
Hb_000946_100 |
0.0 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 2 |
Hb_000016_170 |
0.11521531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006573_290 |
0.1183934102 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 4 |
Hb_002876_020 |
0.1211726 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002154_110 |
0.1214863804 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 6 |
Hb_002677_020 |
0.1226267698 |
- |
- |
PREDICTED: uncharacterized protein LOC105642265 [Jatropha curcas] |
| 7 |
Hb_005496_090 |
0.1251749807 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_001754_080 |
0.126498768 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 9 |
Hb_002320_080 |
0.1272789392 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005167_030 |
0.1308481725 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 11 |
Hb_000928_060 |
0.1309363637 |
- |
- |
transporter, putative [Ricinus communis] |
| 12 |
Hb_145880_020 |
0.1318304638 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 13 |
Hb_000077_200 |
0.1340635913 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 14 |
Hb_000377_150 |
0.1372822162 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 15 |
Hb_000436_030 |
0.1373527621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000815_290 |
0.1387013344 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000649_140 |
0.1405221406 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000771_060 |
0.1405706229 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 19 |
Hb_003106_100 |
0.1415434339 |
- |
- |
mannose-1-phosphate guanyltransferase, putative [Ricinus communis] |
| 20 |
Hb_004007_110 |
0.143181726 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |