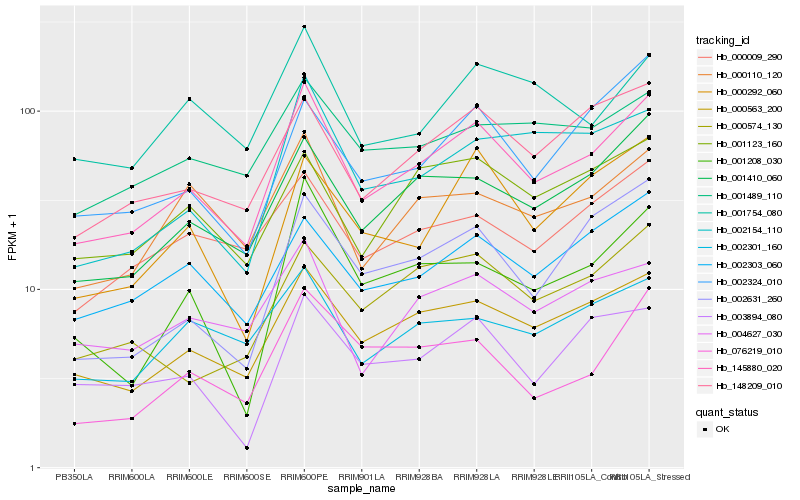
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145880_020 |
0.0 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 2 |
Hb_000563_200 |
0.0819178941 |
- |
- |
hypothetical protein JCGZ_18930 [Jatropha curcas] |
| 3 |
Hb_001410_060 |
0.0947229847 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 4 |
Hb_148209_010 |
0.103810254 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 5 |
Hb_000110_120 |
0.1128418887 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 6 |
Hb_002303_060 |
0.1139668086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004627_030 |
0.1148934048 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 8 |
Hb_002154_110 |
0.1171245362 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 9 |
Hb_000009_290 |
0.1192625874 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 10 |
Hb_000292_060 |
0.1194274523 |
- |
- |
transporter, putative [Ricinus communis] |
| 11 |
Hb_001123_160 |
0.119527975 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_002631_260 |
0.1196066678 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 13 |
Hb_001208_030 |
0.1213393548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_076219_010 |
0.1213967114 |
- |
- |
PREDICTED: RNA pseudouridine synthase 4, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001754_080 |
0.1223945948 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 16 |
Hb_002301_160 |
0.1227350555 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 17 |
Hb_003894_080 |
0.1233187872 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Vitis vinifera] |
| 18 |
Hb_001489_110 |
0.1243604968 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |
| 19 |
Hb_002324_010 |
0.1257465582 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 20 |
Hb_000574_130 |
0.1258384551 |
- |
- |
calcineurin-like protein [Hevea brasiliensis] |