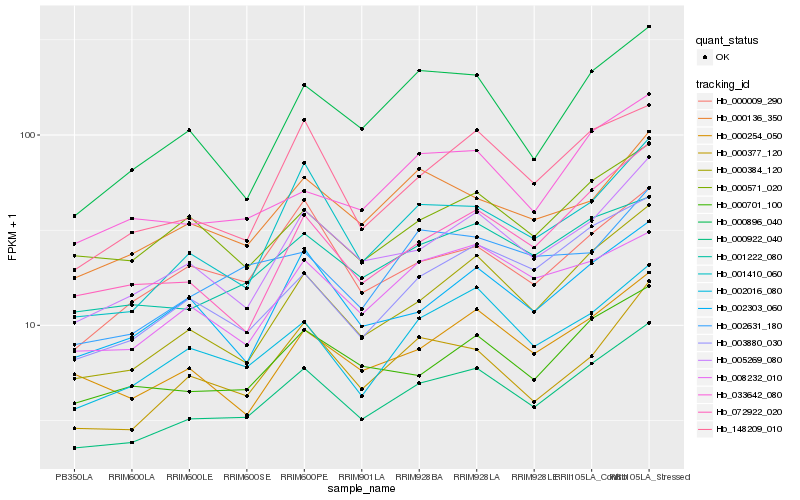
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_040 |
0.0 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 2 |
Hb_005269_080 |
0.065763311 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_000384_120 |
0.0714703597 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001222_080 |
0.0729333262 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002016_080 |
0.0761038312 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000701_100 |
0.0796194284 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 7 |
Hb_001410_060 |
0.0850332406 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 8 |
Hb_148209_010 |
0.0850570062 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 9 |
Hb_003880_030 |
0.0851531955 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000377_120 |
0.0860116469 |
- |
- |
PREDICTED: acyl carrier protein 3, mitochondrial isoform X2 [Jatropha curcas] |
| 11 |
Hb_000254_050 |
0.0874932083 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 12 |
Hb_033642_080 |
0.0876306959 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 13 |
Hb_002631_180 |
0.0898811591 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 14 |
Hb_000136_350 |
0.090605117 |
- |
- |
PREDICTED: 40S ribosomal protein S5 [Nomascus leucogenys] |
| 15 |
Hb_000571_020 |
0.0906414657 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002303_060 |
0.0907459229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008232_010 |
0.0912684523 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000009_290 |
0.0916161645 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 4-like [Jatropha curcas] |
| 19 |
Hb_072922_020 |
0.0926400795 |
- |
- |
SNARE-like superfamily protein [Theobroma cacao] |
| 20 |
Hb_000896_040 |
0.0930914993 |
- |
- |
BnaC02g13240D [Brassica napus] |