| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
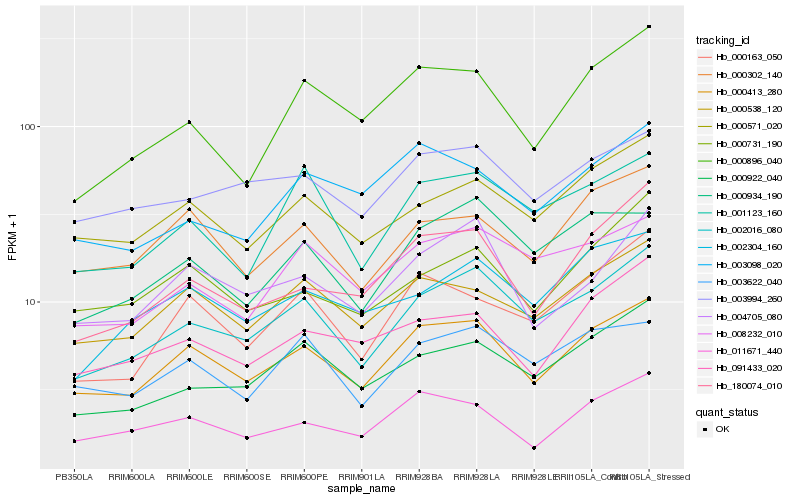
| 1 |
Hb_000413_280 |
0.0 |
- |
- |
PREDICTED: probable UDP-3-O-acylglucosamine N-acyltransferase 2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000538_120 |
0.0721014636 |
transcription factor |
TF Family: SET |
PREDICTED: protein SET DOMAIN GROUP 40 [Jatropha curcas] |
| 3 |
Hb_011671_440 |
0.0736159064 |
- |
- |
ATP-dependent DNA helicase pcrA, putative [Ricinus communis] |
| 4 |
Hb_002016_080 |
0.0772394624 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000302_140 |
0.0805822768 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000571_020 |
0.0844158122 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004705_080 |
0.0903879586 |
transcription factor |
TF Family: ARF |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_003994_260 |
0.0906884687 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 9 |
Hb_003622_040 |
0.090940711 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 10 |
Hb_091433_020 |
0.096527844 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_180074_010 |
0.0974342203 |
- |
- |
Single-stranded DNA-binding protein, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_000163_050 |
0.0985597627 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 13 |
Hb_000896_040 |
0.1001642896 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 14 |
Hb_000922_040 |
0.1010041594 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 15 |
Hb_008232_010 |
0.1016183564 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 16 |
Hb_003098_020 |
0.1021293469 |
- |
- |
40S ribosomal protein S5B [Hevea brasiliensis] |
| 17 |
Hb_000934_190 |
0.102700392 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 18 |
Hb_000731_190 |
0.1029581865 |
- |
- |
PREDICTED: cardiolipin synthase (CMP-forming), mitochondrial [Jatropha curcas] |
| 19 |
Hb_001123_160 |
0.1041030894 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 20 |
Hb_002304_160 |
0.1046015759 |
- |
- |
PREDICTED: nudix hydrolase 15, mitochondrial-like isoform X1 [Jatropha curcas] |