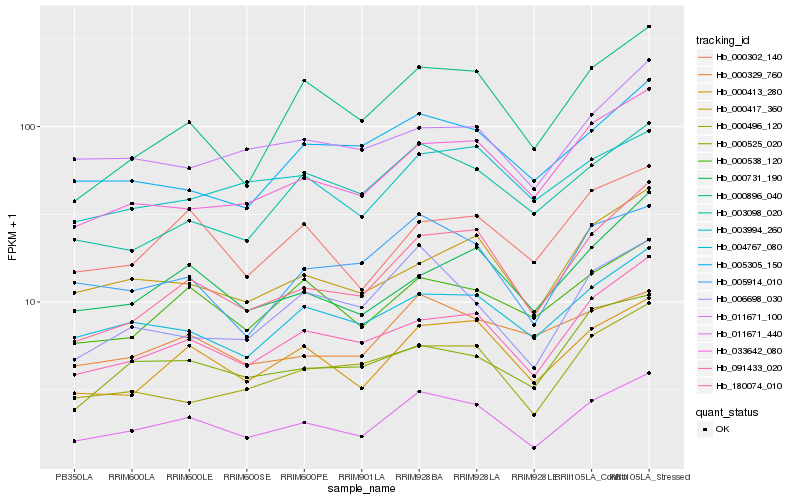
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_440 |
0.0 |
- |
- |
ATP-dependent DNA helicase pcrA, putative [Ricinus communis] |
| 2 |
Hb_000413_280 |
0.0736159064 |
- |
- |
PREDICTED: probable UDP-3-O-acylglucosamine N-acyltransferase 2, mitochondrial [Jatropha curcas] |
| 3 |
Hb_091433_020 |
0.0839852556 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_005914_010 |
0.090711519 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 5 |
Hb_004767_080 |
0.0922427923 |
- |
- |
PREDICTED: mitochondrial ATPase complex subunit ATP10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_180074_010 |
0.092461406 |
- |
- |
Single-stranded DNA-binding protein, mitochondrial precursor, putative [Ricinus communis] |
| 7 |
Hb_000538_120 |
0.0977533927 |
transcription factor |
TF Family: SET |
PREDICTED: protein SET DOMAIN GROUP 40 [Jatropha curcas] |
| 8 |
Hb_033642_080 |
0.0985406523 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 9 |
Hb_000417_360 |
0.1005424548 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000302_140 |
0.1008879162 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 11 |
Hb_006698_030 |
0.1038128492 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 12 |
Hb_000731_190 |
0.1044065817 |
- |
- |
PREDICTED: cardiolipin synthase (CMP-forming), mitochondrial [Jatropha curcas] |
| 13 |
Hb_000496_120 |
0.1062131499 |
- |
- |
PREDICTED: iron-sulfur protein NUBPL [Jatropha curcas] |
| 14 |
Hb_000329_760 |
0.1064193723 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 15 |
Hb_005305_150 |
0.1074505318 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 16 |
Hb_003098_020 |
0.1082128172 |
- |
- |
40S ribosomal protein S5B [Hevea brasiliensis] |
| 17 |
Hb_000896_040 |
0.1098962764 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 18 |
Hb_003994_260 |
0.1108054886 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 19 |
Hb_000525_020 |
0.1113424312 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_011671_100 |
0.1114011393 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |