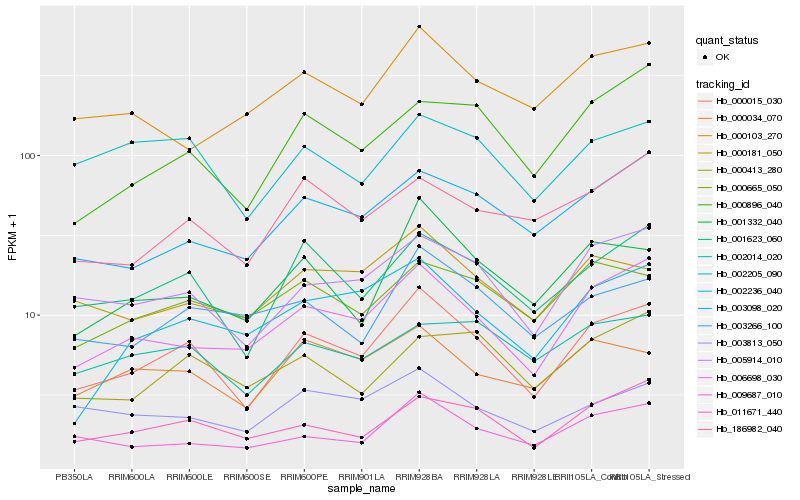
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000015_030 |
0.0 |
- |
- |
PREDICTED: THO complex subunit 4B-like [Jatropha curcas] |
| 2 |
Hb_001623_060 |
0.0930439731 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 3 |
Hb_005914_010 |
0.0955875298 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 4 |
Hb_006698_030 |
0.1001810225 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 5 |
Hb_011671_440 |
0.1134492609 |
- |
- |
ATP-dependent DNA helicase pcrA, putative [Ricinus communis] |
| 6 |
Hb_009687_010 |
0.1173636552 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 7 |
Hb_001332_040 |
0.1177095137 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 8 |
Hb_003098_020 |
0.1195984342 |
- |
- |
40S ribosomal protein S5B [Hevea brasiliensis] |
| 9 |
Hb_000181_050 |
0.126465434 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 10 |
Hb_000896_040 |
0.1282019419 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 11 |
Hb_003266_100 |
0.1288413389 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 12 |
Hb_002236_040 |
0.1291327118 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 13 |
Hb_000034_070 |
0.1298369077 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 3B-like [Jatropha curcas] |
| 14 |
Hb_186982_040 |
0.1324970984 |
- |
- |
hypothetical protein PHAVU_003G089200g [Phaseolus vulgaris] |
| 15 |
Hb_000665_050 |
0.1337154236 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |
| 16 |
Hb_003813_050 |
0.1338599207 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002205_090 |
0.1351406577 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 18 |
Hb_002014_020 |
0.1376639553 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000103_270 |
0.1382261287 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_000413_280 |
0.1386016785 |
- |
- |
PREDICTED: probable UDP-3-O-acylglucosamine N-acyltransferase 2, mitochondrial [Jatropha curcas] |