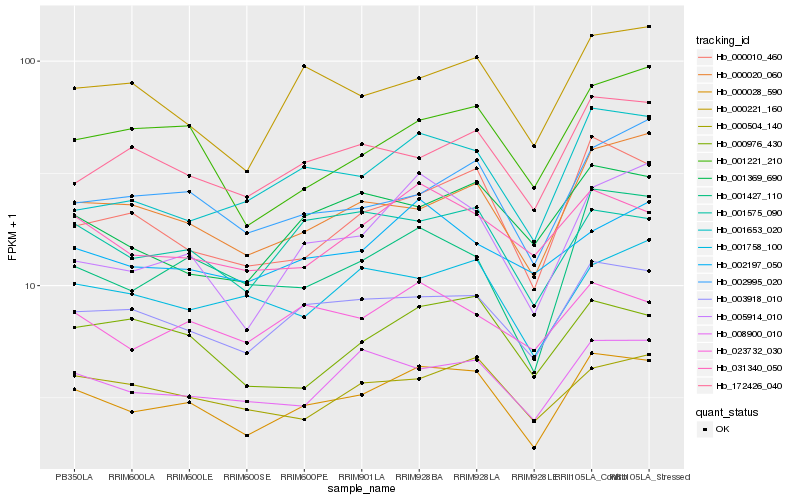
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_590 |
0.0 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 2 |
Hb_003918_010 |
0.0712978984 |
- |
- |
PREDICTED: biotin--protein ligase 2-like [Jatropha curcas] |
| 3 |
Hb_000010_460 |
0.0841255101 |
- |
- |
hypothetical protein EUGRSUZ_B001502, partial [Eucalyptus grandis] |
| 4 |
Hb_008900_010 |
0.0846132906 |
- |
- |
PREDICTED: uncharacterized protein LOC105773666 [Gossypium raimondii] |
| 5 |
Hb_001427_110 |
0.0863409263 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: yrdC domain-containing protein, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000020_060 |
0.0875781034 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 [Jatropha curcas] |
| 7 |
Hb_005914_010 |
0.0884322281 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 8 |
Hb_001575_090 |
0.0887434417 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 9 |
Hb_001653_020 |
0.0897125342 |
transcription factor |
TF Family: C2H2 |
rar1, putative [Ricinus communis] |
| 10 |
Hb_031340_050 |
0.0906022529 |
- |
- |
PREDICTED: tropinone reductase 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000221_160 |
0.0908565135 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6b-1 [Jatropha curcas] |
| 12 |
Hb_002995_020 |
0.0926945821 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 2 [Jatropha curcas] |
| 13 |
Hb_000504_140 |
0.0942472933 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 26, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_172426_040 |
0.0957278428 |
- |
- |
PREDICTED: ELL-associated factor 1 [Jatropha curcas] |
| 15 |
Hb_001221_210 |
0.096728565 |
- |
- |
thiF family protein [Populus trichocarpa] |
| 16 |
Hb_023732_030 |
0.0971939686 |
- |
- |
PREDICTED: proteinaceous RNase P 2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001369_690 |
0.0975890934 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 18 |
Hb_001758_100 |
0.0989613983 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000976_430 |
0.0992914073 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJD5 [Jatropha curcas] |
| 20 |
Hb_002197_050 |
0.1000610511 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |