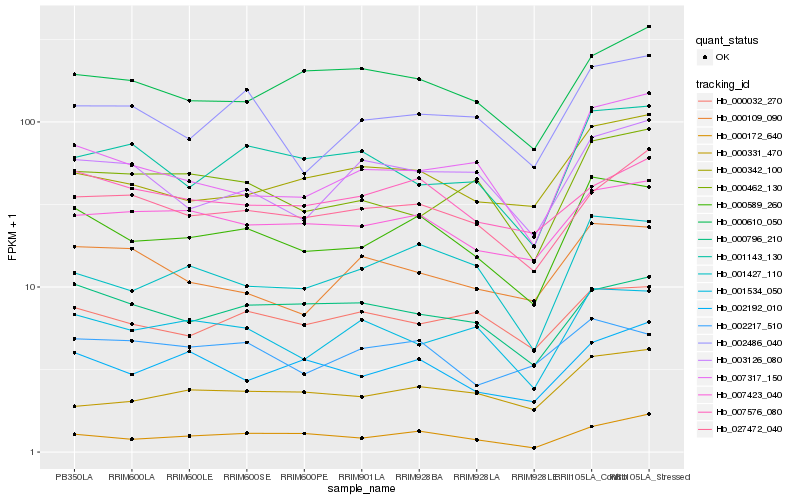
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000589_260 |
0.0 |
- |
- |
3-hydroxybutyryl-CoA dehydratase, putative [Ricinus communis] |
| 2 |
Hb_001427_110 |
0.0918695757 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: yrdC domain-containing protein, mitochondrial [Jatropha curcas] |
| 3 |
Hb_007423_040 |
0.1010338523 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001534_050 |
0.1033059719 |
- |
- |
PREDICTED: uncharacterized protein LOC105642299 [Jatropha curcas] |
| 5 |
Hb_000172_640 |
0.1041579635 |
- |
- |
hypothetical protein POPTR_0007s06250g [Populus trichocarpa] |
| 6 |
Hb_002192_010 |
0.1074034374 |
- |
- |
- |
| 7 |
Hb_002486_040 |
0.107535033 |
- |
- |
- |
| 8 |
Hb_000342_100 |
0.1090480413 |
- |
- |
PREDICTED: PITH domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_007317_150 |
0.1102348511 |
- |
- |
PREDICTED: 40S ribosomal protein S25 [Jatropha curcas] |
| 10 |
Hb_000796_210 |
0.1139043607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_027472_040 |
0.1158704855 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 12 |
Hb_000462_130 |
0.1161483072 |
- |
- |
hypothetical protein CICLE_v10009355mg [Citrus clementina] |
| 13 |
Hb_000032_270 |
0.1162369044 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 13 [Jatropha curcas] |
| 14 |
Hb_003126_080 |
0.1180681785 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 15 |
Hb_001143_130 |
0.1186116159 |
- |
- |
peroxidase 2 [Hevea brasiliensis] |
| 16 |
Hb_000109_090 |
0.1195076848 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 17 |
Hb_002217_510 |
0.1200329771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000610_050 |
0.1214687307 |
- |
- |
skp1, putative [Ricinus communis] |
| 19 |
Hb_007576_080 |
0.1217568205 |
- |
- |
PREDICTED: uncharacterized protein LOC105643972 [Jatropha curcas] |
| 20 |
Hb_000331_470 |
0.1221474001 |
- |
- |
PREDICTED: uncharacterized protein LOC105136689 [Populus euphratica] |