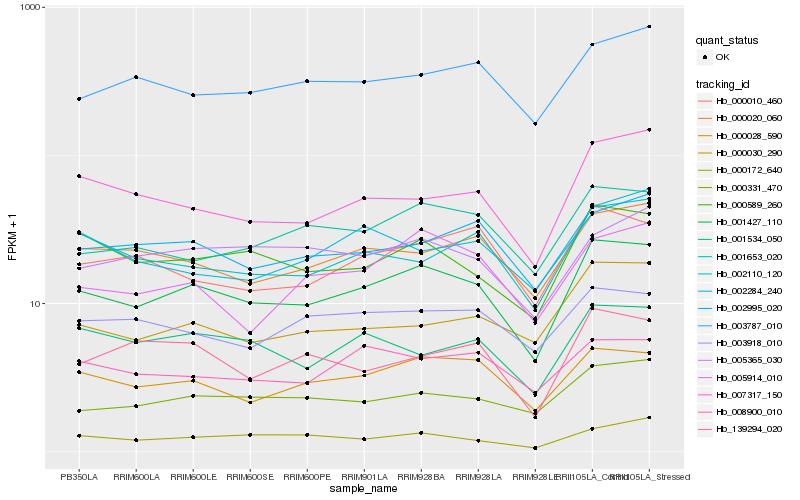
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_110 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: yrdC domain-containing protein, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000028_590 |
0.0863409263 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 3 |
Hb_000589_260 |
0.0918695757 |
- |
- |
3-hydroxybutyryl-CoA dehydratase, putative [Ricinus communis] |
| 4 |
Hb_000020_060 |
0.0978992516 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 [Jatropha curcas] |
| 5 |
Hb_000331_470 |
0.0996432808 |
- |
- |
PREDICTED: uncharacterized protein LOC105136689 [Populus euphratica] |
| 6 |
Hb_001653_020 |
0.1007351635 |
transcription factor |
TF Family: C2H2 |
rar1, putative [Ricinus communis] |
| 7 |
Hb_002995_020 |
0.1029997519 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 2 [Jatropha curcas] |
| 8 |
Hb_003787_010 |
0.1073450516 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Phoenix dactylifera] |
| 9 |
Hb_007317_150 |
0.1075408325 |
- |
- |
PREDICTED: 40S ribosomal protein S25 [Jatropha curcas] |
| 10 |
Hb_000010_460 |
0.1089763106 |
- |
- |
hypothetical protein EUGRSUZ_B001502, partial [Eucalyptus grandis] |
| 11 |
Hb_001534_050 |
0.1102096495 |
- |
- |
PREDICTED: uncharacterized protein LOC105642299 [Jatropha curcas] |
| 12 |
Hb_008900_010 |
0.1115327255 |
- |
- |
PREDICTED: uncharacterized protein LOC105773666 [Gossypium raimondii] |
| 13 |
Hb_000172_640 |
0.1126453259 |
- |
- |
hypothetical protein POPTR_0007s06250g [Populus trichocarpa] |
| 14 |
Hb_005365_030 |
0.1128560224 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 15 |
Hb_003918_010 |
0.1130729203 |
- |
- |
PREDICTED: biotin--protein ligase 2-like [Jatropha curcas] |
| 16 |
Hb_000030_290 |
0.1136070857 |
- |
- |
PREDICTED: uncharacterized protein LOC100247068 [Vitis vinifera] |
| 17 |
Hb_139294_020 |
0.114891331 |
- |
- |
vat protein [Cucumis melo] |
| 18 |
Hb_005914_010 |
0.1155625264 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 19 |
Hb_002110_120 |
0.1159997306 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 20 |
Hb_002284_240 |
0.1160431624 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 115 isoform X3 [Jatropha curcas] |