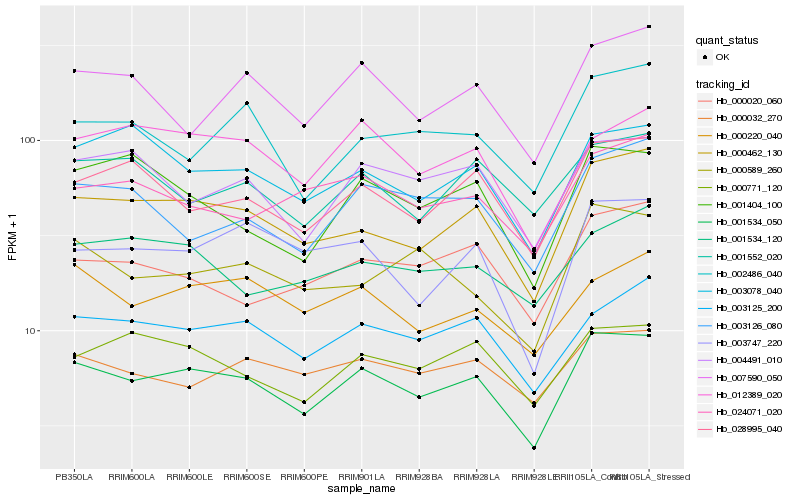
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001534_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642299 [Jatropha curcas] |
| 2 |
Hb_000462_130 |
0.0652810469 |
- |
- |
hypothetical protein CICLE_v10009355mg [Citrus clementina] |
| 3 |
Hb_003125_200 |
0.0826436727 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 4 |
Hb_028995_040 |
0.0863329335 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004491_010 |
0.093772 |
- |
- |
60S ribosomal protein L51, putative [Ricinus communis] |
| 6 |
Hb_000771_120 |
0.0937966317 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003078_040 |
0.0951989776 |
- |
- |
PREDICTED: hit family protein 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003747_220 |
0.0963957785 |
- |
- |
PREDICTED: syntaxin-22-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000220_040 |
0.0967156257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000032_270 |
0.0971923927 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 13 [Jatropha curcas] |
| 11 |
Hb_012389_020 |
0.0973390041 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 12 |
Hb_003126_080 |
0.0974712939 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 13 |
Hb_007590_050 |
0.0976892399 |
- |
- |
NudC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001404_100 |
0.09771602 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K [Gossypium raimondii] |
| 15 |
Hb_000020_060 |
0.09987943 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 [Jatropha curcas] |
| 16 |
Hb_024071_020 |
0.1019309366 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 17 |
Hb_001534_120 |
0.1028967364 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 18 |
Hb_001552_020 |
0.1030183049 |
- |
- |
PREDICTED: uncharacterized protein LOC105639653 [Jatropha curcas] |
| 19 |
Hb_000589_260 |
0.1033059719 |
- |
- |
3-hydroxybutyryl-CoA dehydratase, putative [Ricinus communis] |
| 20 |
Hb_002486_040 |
0.1033707024 |
- |
- |
- |