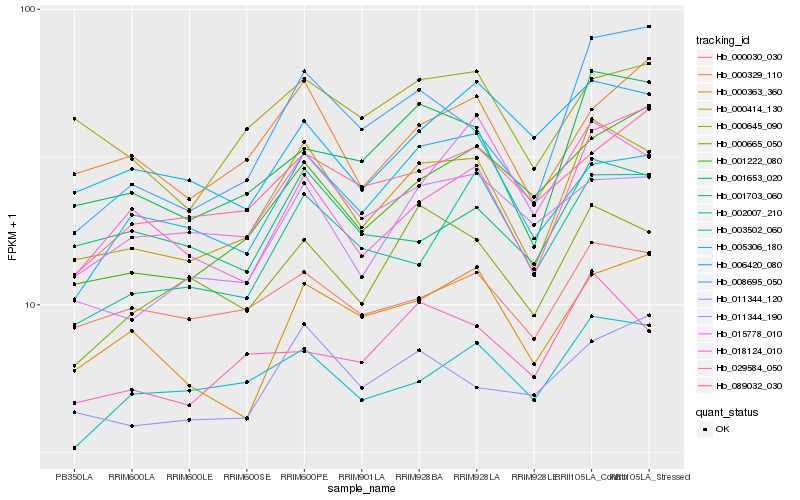
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000414_130 |
0.0 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 2 |
Hb_001653_020 |
0.0761680962 |
transcription factor |
TF Family: C2H2 |
rar1, putative [Ricinus communis] |
| 3 |
Hb_015778_010 |
0.0791511396 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 1 [Jatropha curcas] |
| 4 |
Hb_000665_050 |
0.0816188816 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |
| 5 |
Hb_000030_030 |
0.0901900843 |
- |
- |
PREDICTED: ADP-glucose phosphorylase [Jatropha curcas] |
| 6 |
Hb_003502_060 |
0.090495212 |
- |
- |
PREDICTED: alpha-1,3/1,6-mannosyltransferase ALG2 [Jatropha curcas] |
| 7 |
Hb_005306_180 |
0.0910503004 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_000329_110 |
0.0911125254 |
- |
- |
PREDICTED: uncharacterized protein LOC105643142 [Jatropha curcas] |
| 9 |
Hb_000645_090 |
0.0915817744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_029584_050 |
0.0916137776 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3 [Jatropha curcas] |
| 11 |
Hb_011344_190 |
0.0922366332 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 12 |
Hb_002007_210 |
0.0930052414 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_011344_120 |
0.09491235 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 14 |
Hb_001703_060 |
0.094999754 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000363_360 |
0.0960234987 |
- |
- |
hypothetical protein PRUPE_ppa009146mg [Prunus persica] |
| 16 |
Hb_008695_050 |
0.0962347895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006420_080 |
0.0997930409 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_018124_010 |
0.1021304597 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001222_080 |
0.1026825921 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 20 |
Hb_089032_030 |
0.1029357425 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |