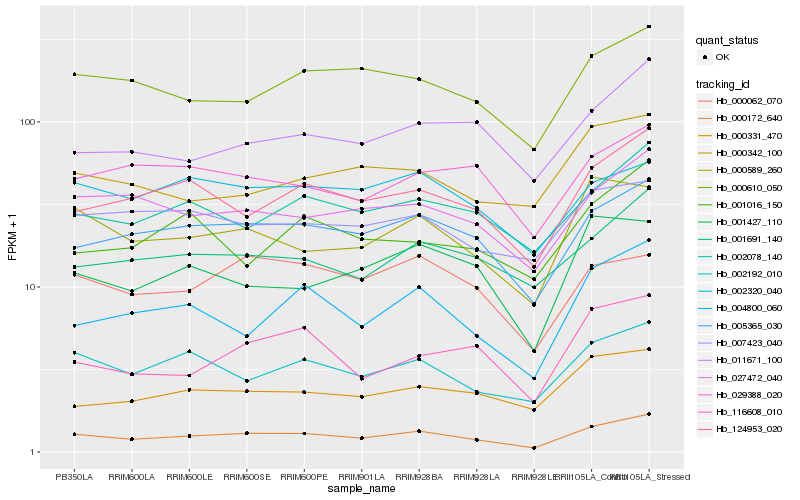
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_640 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s06250g [Populus trichocarpa] |
| 2 |
Hb_005365_030 |
0.0808521409 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 3 |
Hb_124953_020 |
0.0880748054 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 4 |
Hb_004800_060 |
0.0951477736 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 5 |
Hb_002192_010 |
0.0952206881 |
- |
- |
- |
| 6 |
Hb_027472_040 |
0.0972974937 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 7 |
Hb_002078_140 |
0.0976099418 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 8 |
Hb_000610_050 |
0.1000931566 |
- |
- |
skp1, putative [Ricinus communis] |
| 9 |
Hb_001691_140 |
0.1017134399 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 10 |
Hb_000589_260 |
0.1041579635 |
- |
- |
3-hydroxybutyryl-CoA dehydratase, putative [Ricinus communis] |
| 11 |
Hb_011671_100 |
0.1090854269 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |
| 12 |
Hb_000331_470 |
0.110174716 |
- |
- |
PREDICTED: uncharacterized protein LOC105136689 [Populus euphratica] |
| 13 |
Hb_001427_110 |
0.1126453259 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: yrdC domain-containing protein, mitochondrial [Jatropha curcas] |
| 14 |
Hb_116608_010 |
0.1127903967 |
transcription factor |
TF Family: NF-YC |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_029388_020 |
0.1164340769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000342_100 |
0.1184143322 |
- |
- |
PREDICTED: PITH domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_007423_040 |
0.1186776672 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001016_150 |
0.1221852637 |
- |
- |
Rop1 [Hevea brasiliensis] |
| 19 |
Hb_000062_070 |
0.1224603818 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 20 |
Hb_002320_040 |
0.1231050371 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |