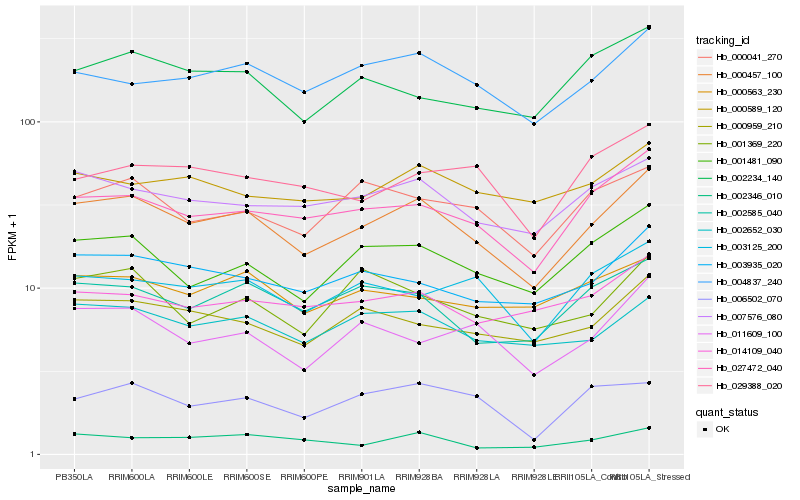
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000457_100 |
0.0 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 2 |
Hb_027472_040 |
0.0760743688 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 3 |
Hb_001481_090 |
0.0851532253 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 4 |
Hb_007576_080 |
0.0860965399 |
- |
- |
PREDICTED: uncharacterized protein LOC105643972 [Jatropha curcas] |
| 5 |
Hb_004837_240 |
0.0862648595 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 6 |
Hb_003935_020 |
0.088293612 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_002585_040 |
0.089988011 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 8 |
Hb_002652_030 |
0.0909203835 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 9 |
Hb_000959_210 |
0.0920971566 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 10 |
Hb_000041_270 |
0.097927266 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 11 |
Hb_003125_200 |
0.0988909181 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 12 |
Hb_011609_100 |
0.1018316182 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_002234_140 |
0.1023618771 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 14 |
Hb_000589_120 |
0.1042620639 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL30 [Gossypium raimondii] |
| 15 |
Hb_014109_040 |
0.1044147074 |
- |
- |
PREDICTED: probable folylpolyglutamate synthase [Jatropha curcas] |
| 16 |
Hb_002346_010 |
0.1045431888 |
- |
- |
- |
| 17 |
Hb_000563_230 |
0.1056082664 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 18 |
Hb_001369_220 |
0.1064476195 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_029388_020 |
0.1075970484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006502_070 |
0.1079190539 |
- |
- |
separase, putative [Ricinus communis] |