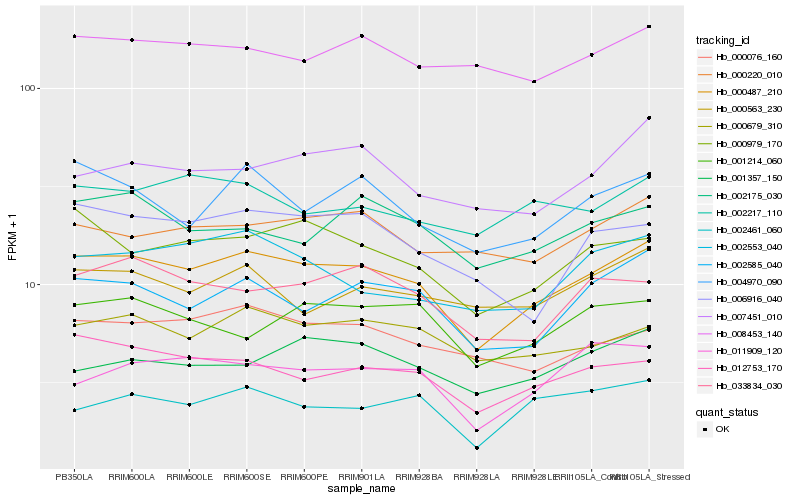
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000487_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 2 |
Hb_002585_040 |
0.0775329303 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 3 |
Hb_012753_170 |
0.0815166306 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000979_170 |
0.0823601287 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 5 |
Hb_000679_310 |
0.0850842829 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004970_090 |
0.0870490802 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 7 |
Hb_033834_030 |
0.0881407922 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_002553_040 |
0.0891804891 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 9 |
Hb_000076_160 |
0.0894940327 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 10 |
Hb_007451_010 |
0.0916517917 |
- |
- |
PREDICTED: 60S ribosome subunit biogenesis protein NIP7 homolog [Jatropha curcas] |
| 11 |
Hb_000220_010 |
0.0922367296 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |
| 12 |
Hb_006916_040 |
0.0929297349 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 13 |
Hb_011909_120 |
0.0942948947 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 14 |
Hb_002461_060 |
0.0950625861 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 15 |
Hb_001214_060 |
0.0950956405 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC8 [Jatropha curcas] |
| 16 |
Hb_002175_030 |
0.0951069679 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 17 |
Hb_002217_110 |
0.0958420146 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 18 |
Hb_000563_230 |
0.0964545757 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 19 |
Hb_001357_150 |
0.0967981748 |
- |
- |
hypothetical protein POPTR_0011s08930g [Populus trichocarpa] |
| 20 |
Hb_008453_140 |
0.0969805023 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |