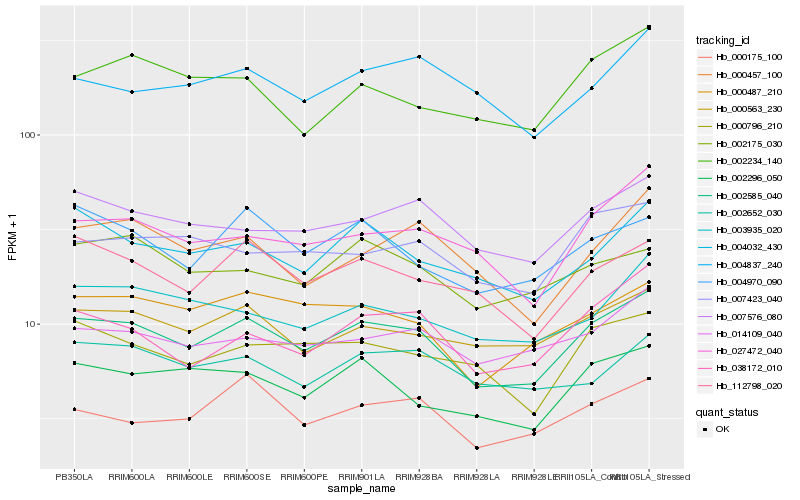
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002585_040 |
0.0 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 2 |
Hb_007576_080 |
0.0753693829 |
- |
- |
PREDICTED: uncharacterized protein LOC105643972 [Jatropha curcas] |
| 3 |
Hb_000563_230 |
0.0761718074 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 4 |
Hb_000487_210 |
0.0775329303 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 5 |
Hb_112798_020 |
0.0795517196 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 6 |
Hb_002296_050 |
0.080098577 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 7 |
Hb_004032_430 |
0.0820743273 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 8 |
Hb_027472_040 |
0.0820989249 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 9 |
Hb_014109_040 |
0.0821809626 |
- |
- |
PREDICTED: probable folylpolyglutamate synthase [Jatropha curcas] |
| 10 |
Hb_000796_210 |
0.0824435113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_038172_010 |
0.0830063338 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 12 |
Hb_000175_100 |
0.0850488453 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 13 |
Hb_003935_020 |
0.0852453957 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_004970_090 |
0.0858712125 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 15 |
Hb_007423_040 |
0.086571773 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002652_030 |
0.0898382567 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 17 |
Hb_000457_100 |
0.089988011 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 18 |
Hb_002234_140 |
0.0911103118 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 19 |
Hb_004837_240 |
0.0913047206 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 20 |
Hb_002175_030 |
0.0926450779 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |