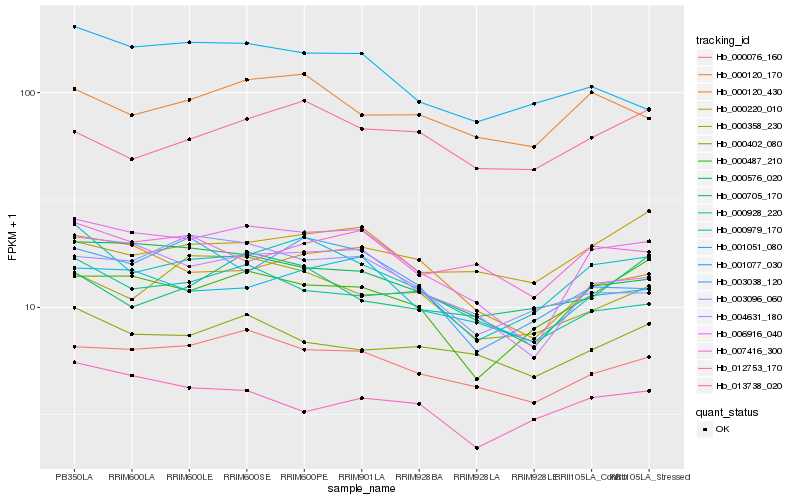
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000979_170 |
0.0 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 2 |
Hb_006916_040 |
0.079992284 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 3 |
Hb_000487_210 |
0.0823601287 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 4 |
Hb_003038_120 |
0.0832901985 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 5 |
Hb_000120_170 |
0.0843492426 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 6 |
Hb_000358_230 |
0.0901994314 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 7 |
Hb_004631_180 |
0.0906197263 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 8 |
Hb_001051_080 |
0.0914065366 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 9 |
Hb_000076_160 |
0.0924461128 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 10 |
Hb_000928_220 |
0.0941531299 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 11 |
Hb_012753_170 |
0.0942929434 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000576_020 |
0.0950153093 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 13 |
Hb_013738_020 |
0.097190652 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 14 |
Hb_000402_080 |
0.0977401668 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 15 |
Hb_000120_430 |
0.0980456429 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 16 |
Hb_007416_300 |
0.0992336477 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 17 |
Hb_001077_030 |
0.0993139472 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000705_170 |
0.0999010956 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 19 |
Hb_003096_060 |
0.1018659941 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 20 |
Hb_000220_010 |
0.1029274462 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |