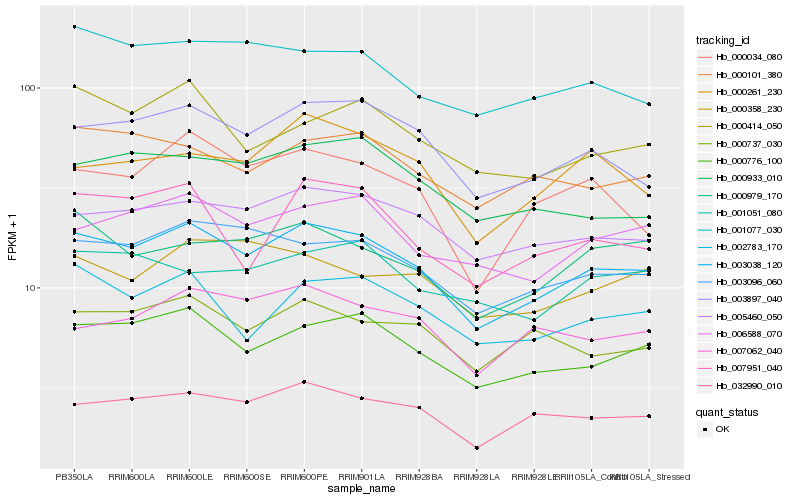
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_120 |
0.0 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 2 |
Hb_000776_100 |
0.0586149506 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 3 |
Hb_003897_040 |
0.0655448578 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 4 |
Hb_032990_010 |
0.0658858607 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 5 |
Hb_003096_060 |
0.0661935966 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 6 |
Hb_005460_050 |
0.0699187193 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 7 |
Hb_006588_070 |
0.0767710192 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 8 |
Hb_002783_170 |
0.07942687 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 9 |
Hb_000737_030 |
0.0799557599 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000034_080 |
0.0806543425 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000101_380 |
0.081103604 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 12 |
Hb_001077_030 |
0.0822346348 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000979_170 |
0.0832901985 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 14 |
Hb_000414_050 |
0.0841651029 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 15 |
Hb_000933_010 |
0.0847597804 |
- |
- |
- |
| 16 |
Hb_007951_040 |
0.0856209009 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 17 |
Hb_007062_040 |
0.0862686457 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 18 |
Hb_000261_230 |
0.0870284478 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 19 |
Hb_001051_080 |
0.0901501236 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 20 |
Hb_000358_230 |
0.0903243211 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |