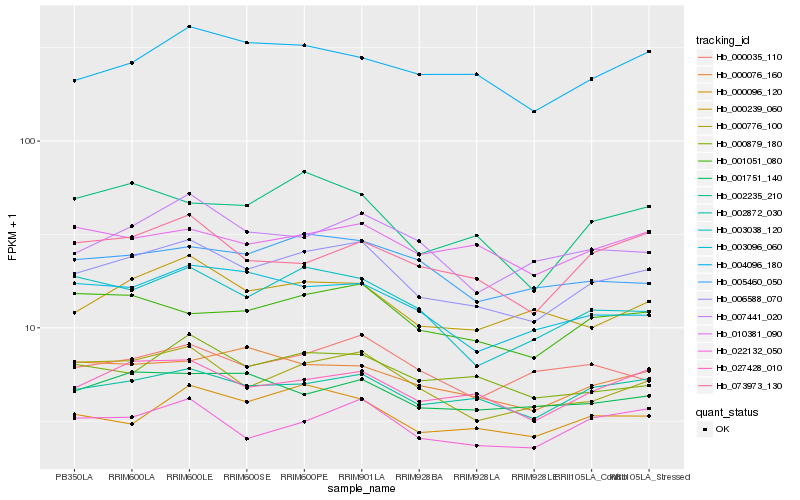
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006588_070 |
0.0 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 2 |
Hb_002872_030 |
0.0606870185 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 3 |
Hb_000776_100 |
0.0620534661 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 4 |
Hb_027428_010 |
0.0631632618 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_022132_050 |
0.0702329434 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 6 |
Hb_001051_080 |
0.0728799508 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 7 |
Hb_000096_120 |
0.0740412039 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 8 |
Hb_000239_060 |
0.0750923215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003038_120 |
0.0767710192 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 10 |
Hb_000879_180 |
0.0777232904 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 11 |
Hb_007441_020 |
0.0779692942 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 12 |
Hb_005460_050 |
0.078830166 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 13 |
Hb_004096_180 |
0.0788538787 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 14 |
Hb_003096_060 |
0.0801185824 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 15 |
Hb_073973_130 |
0.0811037363 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_000076_160 |
0.0815060084 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 17 |
Hb_000035_110 |
0.0816555242 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 18 |
Hb_001751_140 |
0.081773981 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002235_210 |
0.0820814344 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |
| 20 |
Hb_010381_090 |
0.0833304733 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |