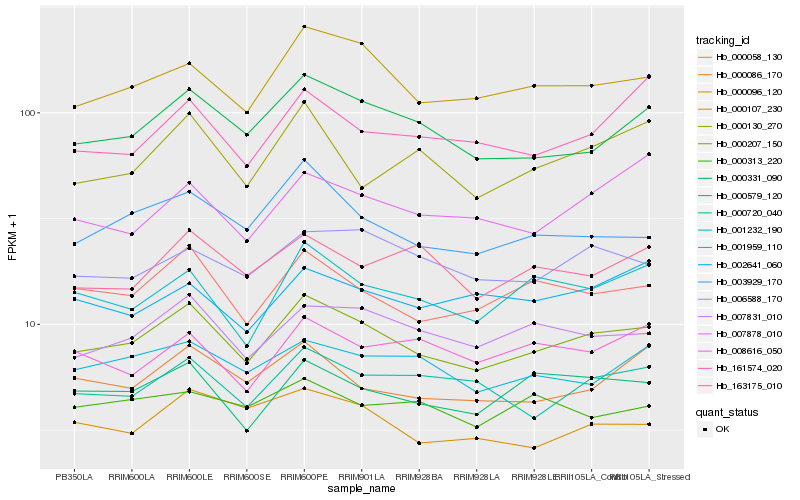
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000207_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 2 |
Hb_000107_230 |
0.0596260391 |
- |
- |
unknown [Medicago truncatula] |
| 3 |
Hb_000058_130 |
0.0619130721 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 4 |
Hb_000331_090 |
0.0623332733 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 5 |
Hb_007831_010 |
0.0676022125 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 6 |
Hb_003929_170 |
0.0696532228 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 7 |
Hb_000579_120 |
0.0705014486 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 8 |
Hb_001232_190 |
0.0735959914 |
- |
- |
PREDICTED: uncharacterized protein LOC105639761 isoform X2 [Jatropha curcas] |
| 9 |
Hb_006588_170 |
0.0756404464 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 10 |
Hb_000720_040 |
0.075785281 |
- |
- |
PREDICTED: replication factor C subunit 2 [Jatropha curcas] |
| 11 |
Hb_001959_110 |
0.0768021444 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000086_170 |
0.0772910551 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 13 |
Hb_000096_120 |
0.0806149582 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 14 |
Hb_163175_010 |
0.0827639625 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 15 |
Hb_000130_270 |
0.0865640932 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002641_060 |
0.088054505 |
- |
- |
prefoldin subunit, putative [Ricinus communis] |
| 17 |
Hb_007878_010 |
0.0883666529 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 4 [Jatropha curcas] |
| 18 |
Hb_008616_050 |
0.0887300412 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 19 |
Hb_161574_020 |
0.0888226834 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 20 |
Hb_000313_220 |
0.0897758196 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |