| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
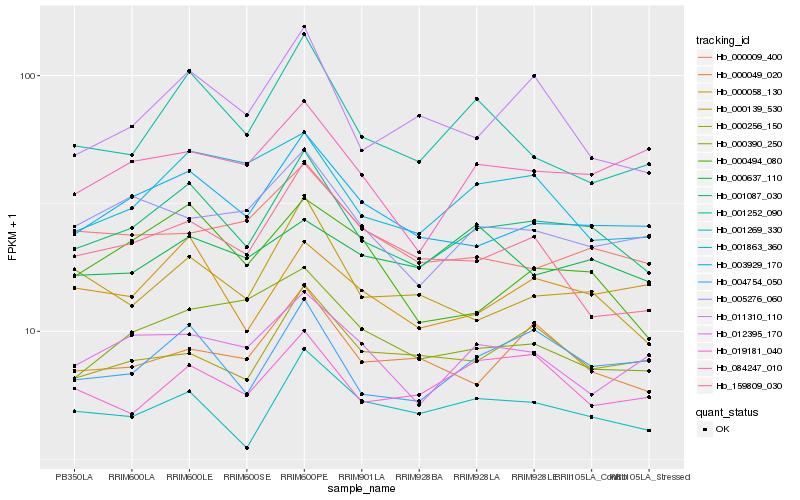
Hb_001087_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 2 |
Hb_003929_170 |
0.0669483952 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 3 |
Hb_004754_050 |
0.0727500452 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 4 |
Hb_001269_330 |
0.0733186228 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 5 |
Hb_000139_530 |
0.0820476424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000058_130 |
0.0861656418 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 7 |
Hb_000049_020 |
0.0870037337 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 8 |
Hb_001863_360 |
0.0878078001 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 9 |
Hb_011310_110 |
0.089127023 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001252_090 |
0.0906253523 |
- |
- |
GDP-D-mannose pyrophosphorylase [Camellia sinensis] |
| 11 |
Hb_000256_150 |
0.0917508032 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 12 |
Hb_005276_060 |
0.0917988575 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_019181_040 |
0.092714613 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 14 |
Hb_000494_080 |
0.092821126 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_012395_170 |
0.0932032542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000390_250 |
0.096038129 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 17 |
Hb_159809_030 |
0.0960496023 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000009_400 |
0.0964552633 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 19 |
Hb_084247_010 |
0.0964849622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000637_110 |
0.0980989255 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |