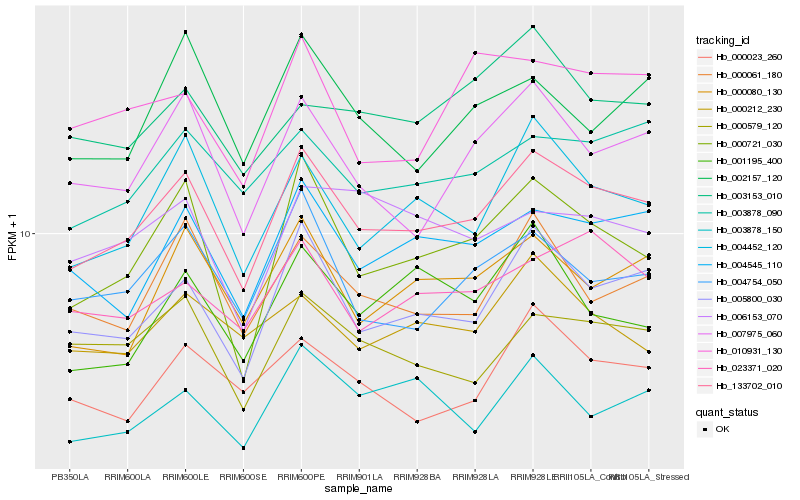
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133702_010 |
0.0 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_005800_030 |
0.0512228688 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 3 |
Hb_004452_120 |
0.0613715127 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 4 |
Hb_004545_110 |
0.0630289354 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_000721_030 |
0.0653297977 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 6 |
Hb_007975_060 |
0.0689633834 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 7 |
Hb_003878_090 |
0.0699713888 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 8 |
Hb_000080_130 |
0.0703372298 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 9 |
Hb_000061_180 |
0.0704669263 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 10 |
Hb_004754_050 |
0.0717365159 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 11 |
Hb_000579_120 |
0.0724494008 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 12 |
Hb_023371_020 |
0.0748229106 |
- |
- |
PREDICTED: uncharacterized protein LOC105633512 [Jatropha curcas] |
| 13 |
Hb_000023_260 |
0.0761592147 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003878_150 |
0.0771452658 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 15 |
Hb_000212_230 |
0.0795990152 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 16 |
Hb_002157_120 |
0.0811903786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003153_010 |
0.0832814111 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 18 |
Hb_001195_400 |
0.0851764099 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 19 |
Hb_010931_130 |
0.0853944059 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 20 |
Hb_006153_070 |
0.0854094266 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |