| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
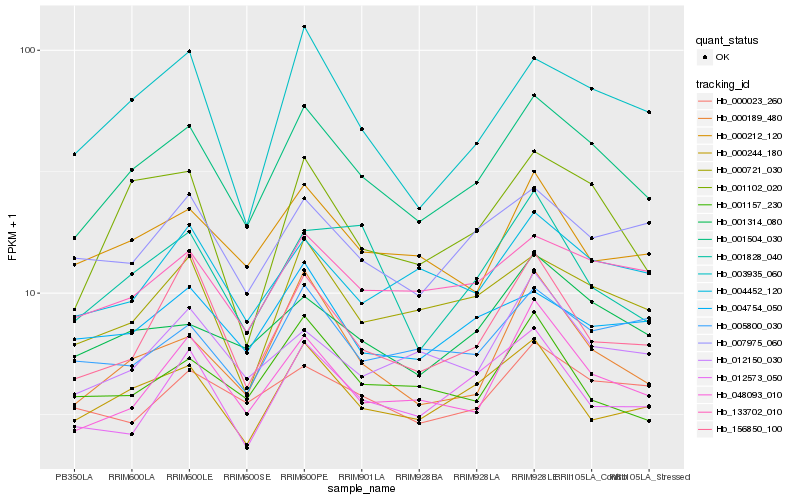
Hb_001504_030 |
0.0 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 2 |
Hb_048093_010 |
0.0807422114 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_000189_480 |
0.0859229179 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 4 |
Hb_133702_010 |
0.0878594411 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_001314_080 |
0.088684399 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 6 |
Hb_000721_030 |
0.0888269356 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 7 |
Hb_001102_020 |
0.0903898782 |
- |
- |
solanesyl diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_000244_180 |
0.0911964308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003935_060 |
0.093232215 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 10 |
Hb_156850_100 |
0.0968247833 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 11 |
Hb_004452_120 |
0.0984399983 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 12 |
Hb_000212_120 |
0.0995491166 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 13 |
Hb_007975_060 |
0.1017991552 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 14 |
Hb_012573_050 |
0.1022740268 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 15 |
Hb_012150_030 |
0.1045849871 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_004754_050 |
0.1056975688 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 17 |
Hb_001828_040 |
0.1061891708 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 18 |
Hb_000023_260 |
0.1067153508 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001157_230 |
0.1082779382 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 20 |
Hb_005800_030 |
0.1091697511 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |