| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
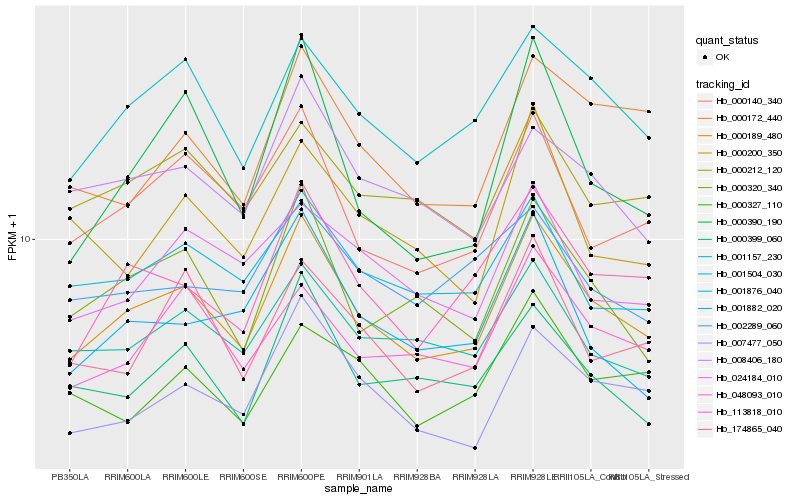
Hb_000189_480 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 2 |
Hb_001504_030 |
0.0859229179 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 3 |
Hb_113818_010 |
0.0871936732 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 4 |
Hb_000320_340 |
0.093579814 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 5 |
Hb_000140_340 |
0.0944340264 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 6 |
Hb_001157_230 |
0.0977458751 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 7 |
Hb_007477_050 |
0.0985502683 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_048093_010 |
0.100410882 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_000399_060 |
0.106192937 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 10 |
Hb_008406_180 |
0.1066639282 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 11 |
Hb_001876_040 |
0.1067950894 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 12 |
Hb_000212_120 |
0.1070757578 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 13 |
Hb_174865_040 |
0.1091742117 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000172_440 |
0.1095594303 |
- |
- |
PREDICTED: NADPH-dependent 1-acyldihydroxyacetone phosphate reductase [Jatropha curcas] |
| 15 |
Hb_024184_010 |
0.111613817 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_000390_190 |
0.1116357938 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_001882_020 |
0.1134371826 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 18 |
Hb_000327_110 |
0.1164835976 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 19 |
Hb_002289_060 |
0.117193483 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000200_350 |
0.1179697257 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |