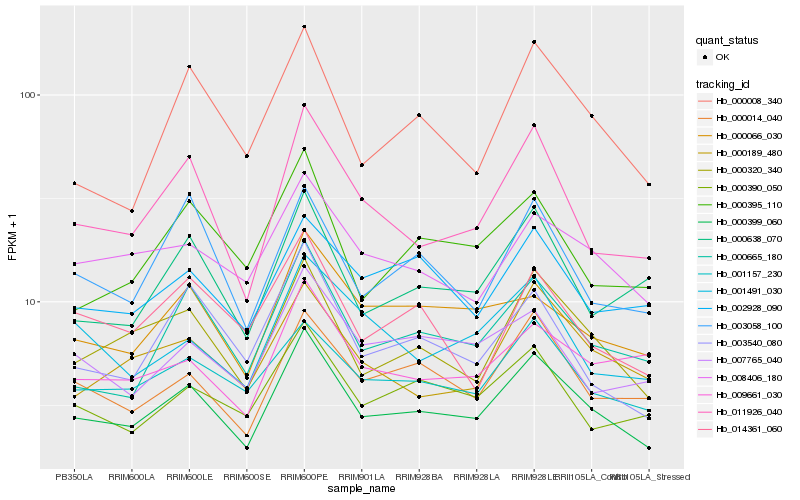
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000399_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 2 |
Hb_000320_340 |
0.0784055577 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 3 |
Hb_011926_040 |
0.0885127657 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000014_040 |
0.0933907017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000665_180 |
0.0947509202 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_001157_230 |
0.0954477513 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 7 |
Hb_008406_180 |
0.0987599736 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_000390_050 |
0.1019560127 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 9 |
Hb_000189_480 |
0.106192937 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 10 |
Hb_003540_080 |
0.1069164957 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000638_070 |
0.1072682768 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 12 |
Hb_000066_030 |
0.1073156797 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007765_040 |
0.1080214507 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 14 |
Hb_000008_340 |
0.1100490162 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 15 |
Hb_002928_090 |
0.1103783971 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 16 |
Hb_009661_030 |
0.1114796345 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_003058_100 |
0.1124470529 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000395_110 |
0.1125053349 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 19 |
Hb_001491_030 |
0.1141858573 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 20 |
Hb_014361_060 |
0.1148623417 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |