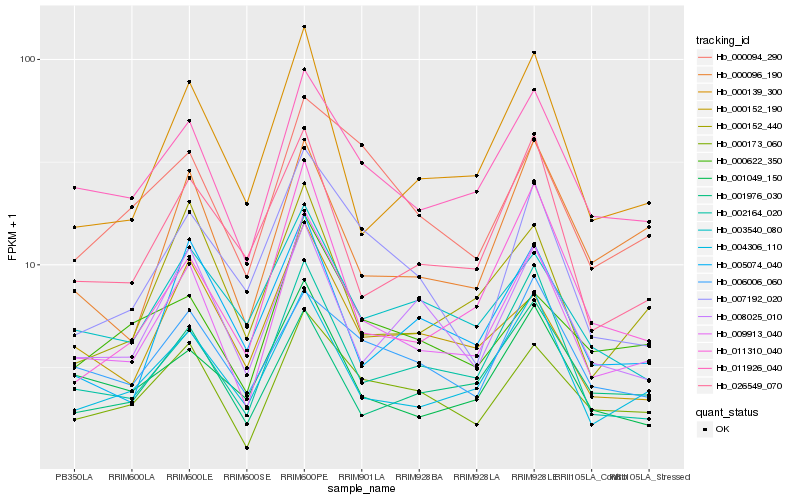
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009913_040 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 2 |
Hb_011926_040 |
0.0838877426 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000173_060 |
0.0906787097 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 4 |
Hb_004306_110 |
0.1065568742 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 5 |
Hb_002164_020 |
0.1104441673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000139_300 |
0.1180461784 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 7 |
Hb_008025_010 |
0.1270571872 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 8 |
Hb_001049_150 |
0.1297351152 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 9 |
Hb_026549_070 |
0.1318230041 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 10 |
Hb_007192_020 |
0.1318476815 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 11 |
Hb_000152_440 |
0.1325849723 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 12 |
Hb_000096_190 |
0.1331245001 |
- |
- |
unknown [Lotus japonicus] |
| 13 |
Hb_000152_190 |
0.1335445293 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |
| 14 |
Hb_000094_290 |
0.1339004662 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_006006_060 |
0.1341493801 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_011310_040 |
0.1365138896 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 17 |
Hb_003540_080 |
0.1369907101 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001976_030 |
0.1370634204 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_005074_040 |
0.1371026974 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000622_350 |
0.1376251385 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |