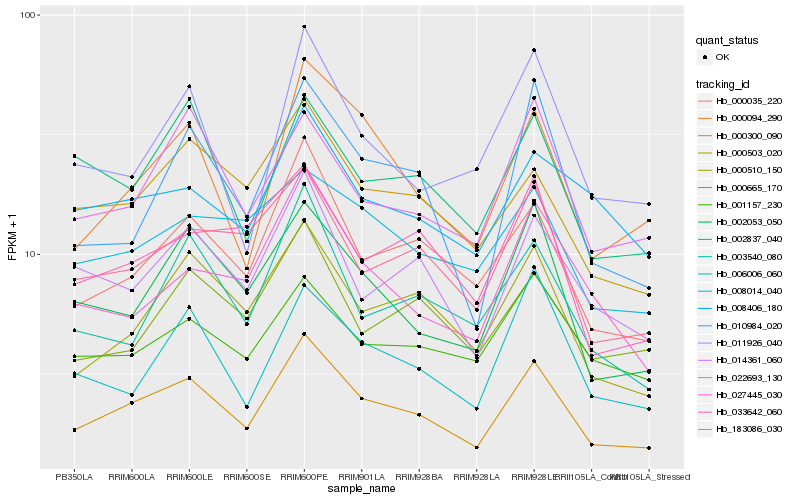
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_090 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_000035_220 |
0.0878960312 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 3 |
Hb_000510_150 |
0.0955446789 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_014361_060 |
0.1001102613 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 5 |
Hb_000094_290 |
0.1005165189 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000503_020 |
0.1008312323 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 7 |
Hb_002053_050 |
0.1017320627 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 8 |
Hb_183086_030 |
0.1039863504 |
- |
- |
GTPase-activating protein GYP7 [Gossypium arboreum] |
| 9 |
Hb_022693_130 |
0.1075876286 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000665_170 |
0.1112955883 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 11 |
Hb_003540_080 |
0.1115504955 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_006006_060 |
0.1124851178 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001157_230 |
0.1125557066 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 14 |
Hb_002837_040 |
0.1125736042 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 15 |
Hb_011926_040 |
0.1146068693 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008406_180 |
0.115058153 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_008014_040 |
0.1158476577 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 18 |
Hb_027445_030 |
0.1160182887 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_033642_060 |
0.1165224983 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_010984_020 |
0.1166238276 |
- |
- |
Translation factor GUF1 like, chloroplastic [Glycine soja] |