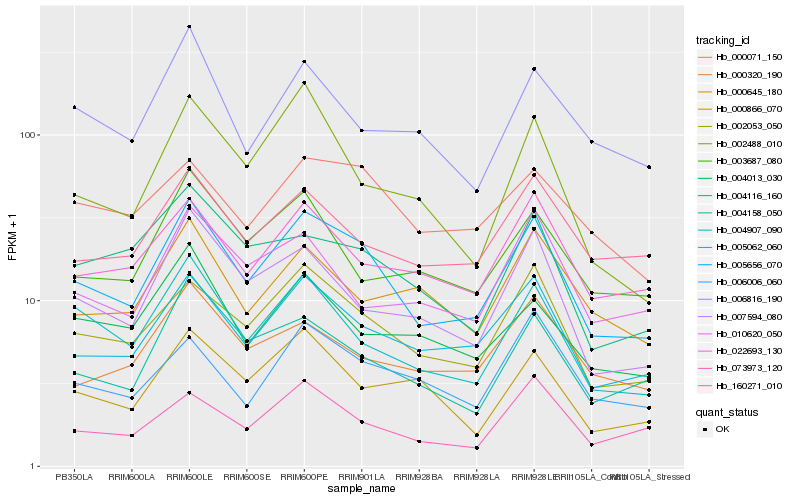
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005656_070 |
0.0 |
- |
- |
PREDICTED: cryptochrome DASH, chloroplastic/mitochondrial [Jatropha curcas] |
| 2 |
Hb_002053_050 |
0.0876703321 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 3 |
Hb_005062_060 |
0.0914215533 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 4 |
Hb_004907_090 |
0.1105681576 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_007594_080 |
0.119188674 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 6 |
Hb_022693_130 |
0.1213529427 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_004158_050 |
0.1232062984 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 8 |
Hb_160271_010 |
0.1286959411 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_073973_120 |
0.1291500921 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000071_150 |
0.1296584162 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 11 |
Hb_000866_070 |
0.1334548781 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 12 |
Hb_003687_080 |
0.1345993388 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 13 |
Hb_006006_060 |
0.1354834669 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006816_190 |
0.1357665986 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 15 |
Hb_000320_190 |
0.1366296517 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_004013_030 |
0.1372335279 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 17 |
Hb_004116_160 |
0.1379141049 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002488_010 |
0.138112062 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 19 |
Hb_010620_050 |
0.1391722421 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 20 |
Hb_000645_180 |
0.1396026892 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |