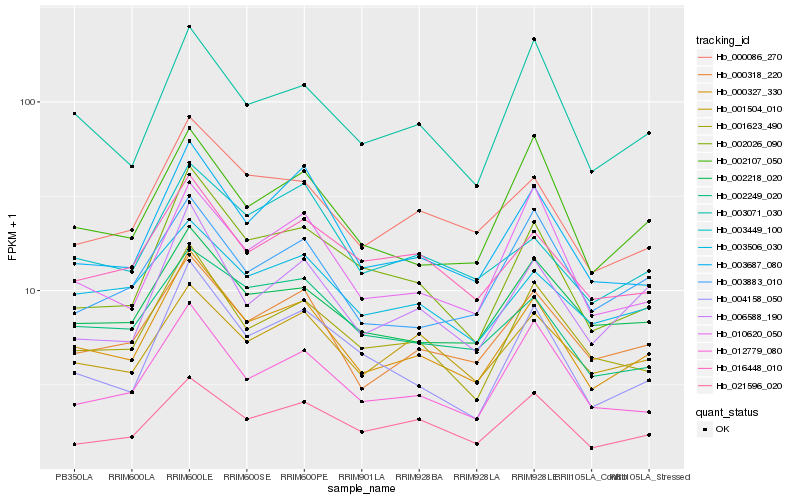
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_330 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000318_220 |
0.0681846521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003687_080 |
0.0782592582 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 4 |
Hb_001623_490 |
0.0791184319 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000086_270 |
0.0803776817 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 6 |
Hb_002218_020 |
0.0826910964 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 7 |
Hb_003071_030 |
0.0839710406 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006588_190 |
0.0847606327 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 9 |
Hb_002026_090 |
0.0859549953 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 10 |
Hb_002107_050 |
0.0891049391 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 11 |
Hb_004158_050 |
0.0892169281 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 12 |
Hb_012779_080 |
0.0894587944 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 13 |
Hb_021596_020 |
0.0929421566 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 14 |
Hb_003506_030 |
0.0932234695 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 15 |
Hb_016448_010 |
0.0940073929 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 16 |
Hb_010620_050 |
0.0943868596 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 17 |
Hb_002249_020 |
0.0945130517 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 18 |
Hb_001504_010 |
0.0952157151 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 19 |
Hb_003449_100 |
0.0961397601 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 20 |
Hb_003883_010 |
0.0993295984 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |