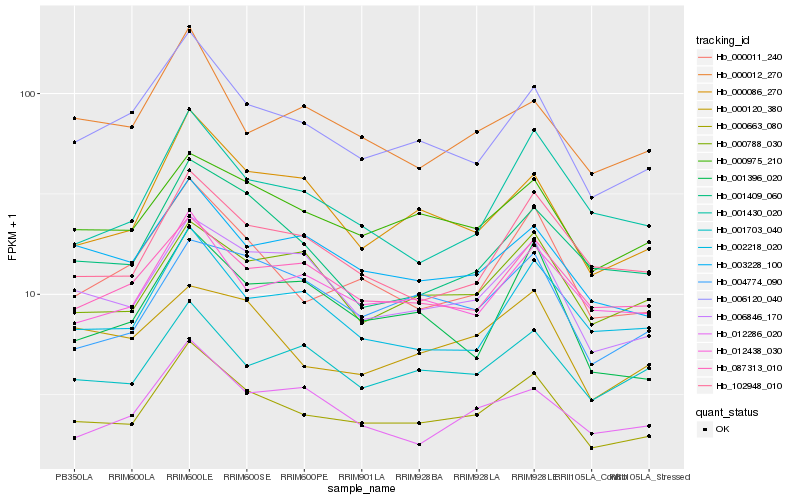
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000663_080 |
0.0 |
transcription factor |
TF Family: HB |
Homeobox protein HAT3.1, putative [Ricinus communis] |
| 2 |
Hb_006120_040 |
0.0731742953 |
- |
- |
PREDICTED: prostatic spermine-binding protein [Jatropha curcas] |
| 3 |
Hb_000011_240 |
0.0740189932 |
- |
- |
PREDICTED: uncharacterized protein LOC105631316 [Jatropha curcas] |
| 4 |
Hb_000086_270 |
0.084221703 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 5 |
Hb_000975_210 |
0.0844433896 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 6 |
Hb_006846_170 |
0.0850374437 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 7 |
Hb_003228_100 |
0.0929012635 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 8 |
Hb_012438_030 |
0.0936418587 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 9 |
Hb_102948_010 |
0.0945457428 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002218_020 |
0.0962459843 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 11 |
Hb_087313_010 |
0.0993658092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001703_040 |
0.1012442218 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001409_060 |
0.10130697 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 14 |
Hb_004774_090 |
0.1032559269 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 15 |
Hb_000788_030 |
0.1040025826 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 16 |
Hb_012286_020 |
0.1040383286 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 17 |
Hb_000012_270 |
0.1056157245 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 18 |
Hb_001396_020 |
0.1056729763 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 19 |
Hb_001430_020 |
0.105959141 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000120_380 |
0.1063955245 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |