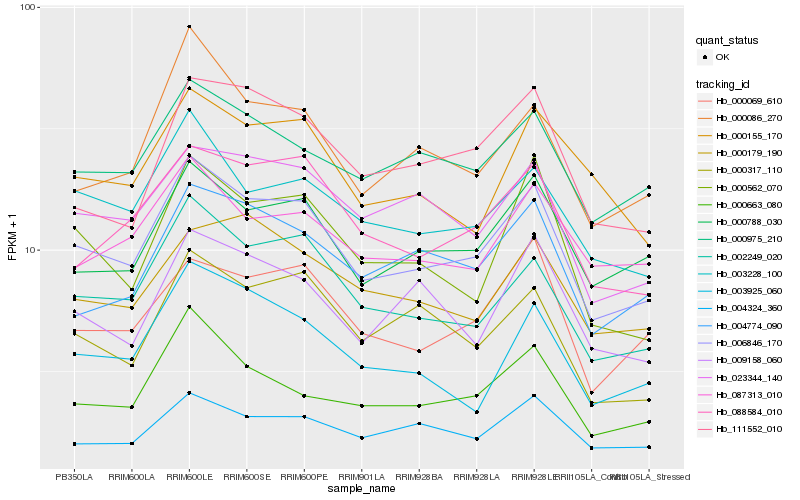
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006846_170 |
0.0 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 2 |
Hb_002249_020 |
0.0645393425 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 3 |
Hb_000788_030 |
0.0736253778 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 4 |
Hb_003228_100 |
0.0783389973 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 5 |
Hb_000975_210 |
0.0817572246 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 6 |
Hb_088584_010 |
0.0822024014 |
- |
- |
hypothetical protein POPTR_0016s08470g [Populus trichocarpa] |
| 7 |
Hb_023344_140 |
0.082947965 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |
| 8 |
Hb_000069_610 |
0.0833044034 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 9 |
Hb_000663_080 |
0.0850374437 |
transcription factor |
TF Family: HB |
Homeobox protein HAT3.1, putative [Ricinus communis] |
| 10 |
Hb_000086_270 |
0.0865420446 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 11 |
Hb_000562_070 |
0.0888671637 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000317_110 |
0.0892947186 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_003925_060 |
0.0902410741 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004774_090 |
0.0910936743 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 15 |
Hb_087313_010 |
0.0919143599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_111552_010 |
0.0922936575 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 17 |
Hb_009158_060 |
0.0923841575 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 18 |
Hb_004324_360 |
0.0931130218 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000179_190 |
0.0932344808 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 20 |
Hb_000155_170 |
0.0957111696 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |