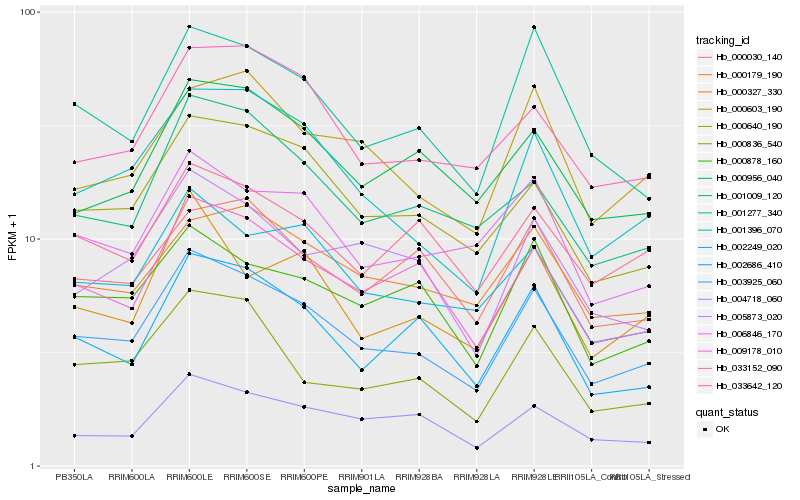
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003925_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009178_010 |
0.0742852141 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 3 |
Hb_001396_070 |
0.0752176019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000640_190 |
0.0772616338 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002249_020 |
0.0772878784 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 6 |
Hb_001277_340 |
0.0846865287 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 7 |
Hb_002686_410 |
0.0878593315 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_033152_090 |
0.0889022387 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 9 |
Hb_006846_170 |
0.0902410741 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 10 |
Hb_001009_120 |
0.0905404269 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000878_160 |
0.0913435927 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 12 |
Hb_000179_190 |
0.094496662 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 13 |
Hb_000836_540 |
0.0955115396 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 14 |
Hb_004718_060 |
0.0955382844 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000956_040 |
0.0956357899 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 16 |
Hb_000603_190 |
0.0956674945 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 17 |
Hb_005873_020 |
0.0960982696 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 18 |
Hb_033642_120 |
0.0988541148 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 19 |
Hb_000030_140 |
0.0992392041 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000327_330 |
0.0998616006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |